A tree kernel to analyze phylogenetic profiles
|
|
|
- Roger Maxwell
- 5 years ago
- Views:
Transcription
1 BIOINFORMATICS Vol. no. 22 Pages 9 A tree kernel to analyze phylogenetic profiles Jean-Philippe Vert Bioinformatics Center, Institute for Chemical Research, Kyoto University, Uji, Kyoto, 6-, Japan ABSTRACT Motivation: The phylogenetic profile of a protein is a string that encodes the presence or absence of the protein in every fully sequenced genome. Because proteins that participate in a common structural complex or metabolic pathway are likely to evolve in a correlated fashion, the phylogenetic profiles of such proteins are often similar or at least related to each other. The question we address in this paper is the following: how to measure the similarity between two profiles, in an evolutionarily relevant way, in order to develop efficient function prediction methods? Results: We show how the profiles can be mapped to a high-dimensional vector space which incorporates evolutionarily relevant information, and we provide an algorithm to compute efficiently the inner product in that space, which we call the tree kernel. The tree kernel can be used by any kernel-based analysis method for classification or data mining of phylogenetic profiles. As an application a Support Vector Machine (SVM) trained to predict the functional class of a gene from its phylogenetic profile is shown to perform better with the tree kernel than with a naive kernel that does not include any information about the phylogenetic relationships among species. Moreover a kernel principal component analysis (KPCA) of the phylogenetic profiles illustrates the sensitivity of the tree kernel to evolutionarily relevant variations. Availability: All data and softwares used are freely and publicly available upon request. Contact: Jean-Philippe.Vert@mines.org INTRODUCTION The availability of an increasing number of fully sequenced genomes has promoted the development of large-scale comparative genomic approaches to better understand the machinery of the cell. One such approach is the use of phylogenetic profiles (Pellegrini et al., 999) to assign functions to genes based on their patterns of inheritance across species. The hypothesis behind this method is that proteins that participate in a common structural complex or metabolic pathway evolve in a correlated fashion, and should therefore be present in the same organisms. As a matter of fact the phylogenetic profile of a gene has been shown to provide information about its function (Pellegrini et al., 999) and is an interesting alternative to other approaches to gene function prediction based on direct sequence comparisons or gene co-expression (Marcotte et al., 999). In its simplest form the phylogenetic profile of a gene is a string of bits, each bit indicating the presence or absence of a homologue of the gene in a different organism. Once the phylogenetic profiles for all the genes of a given organism have been computed, the function of a gene can be inferred to some extent by examining the functions of other genes with similar or at least related profiles. A fundamental question addressed in this paper is therefore the following: how to measure the similarity between phylogenetic profiles? The simplest approach to quantify the difference between two profiles is to count the number of organisms where they differ. Intuitively, the smaller this number, the more similar the profiles. This method was used in (Pellegrini et al., 999) where two profiles are defined as neighbors when they differ by less than 3 bits. A more appealing measure is the differential parsimony introduced in (Liberles et al., 22), based on the phylogenetic reconstruction of the ancestors and the comparison of the reconstructed trees. Indeed, it is intuitively more meaningful to compare the whole historical evolutions of two genes, rather than just their presence or absence in current organisms. In this paper we generalize this idea and embed it into a powerful mathematical framework that opens new analysis opportunities. Instead of just defining a measure of similarity between profiles, such as the number of bits where they differ or the differential parsimony of the reconstructed phylogenetic trees, we propose to map each profile to a high-dimensional vector space (which we call feature space), defined in such a way that each coordinate in the feature space corresponds to one particular pattern of inheritance during evolution. As an example, one pattern of inheritance could be the gene has been transmitted to proteobacteria and eukaryotes, but not to Gram-positive bacteria. As it is impossible to know for sure the ancestor genomes the mapping of a phylogenetic profile to the feature space is defined thanks to a probabilistic model of evolution (a classical Bayesian tree model) to give weights to features that correspond to c Oxford University Press 2
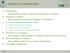
2 J.-P.Vert! plausible patterns of inheritance for the given profile. If one considers all possible patterns of evolution the dimension of the feature space is very large, and the explicit computation of the image of a profile could be prohibitive in terms of computation time and memory storage. However, as a main contribution, this paper provides an algorithm to compute very simply and quickly the inner product between the images of any two profiles in the feature space, which does not involve the explicit computation of each image. The resulting function, which maps any two profiles to the inner product of their images in the high-dimensional feature space, is called a tree kernel. It belongs to a larger class of functions, called kernels, defined as the inner product of two objects mapped to any vector space. Once a kernel and a corresponding feature space are chosen, it is possible to define the distance between two profiles as the Euclidean distance between their images in the feature space. In the case of the tree kernel presented in this paper, two profiles are near each other in the feature space if they are likely to have shared common patterns of inheritance during evolution, which is an appealing property. However, more than just computing a Euclidean distance can be done once a kernel is defined. In particular a whole set of algorithms, known as kernel methods, have been developed recently and shown to be very helpful in many real-world applications (Cristianini & Shawe- Taylor, 2; Schölkopf & Smola, 2). Kernel methods work implicitly in the feature space using only the kernel function, and include such diverse algorithms as Support Vector Machines for classification or regression (Vapnik, 998), kernel principal component analysis (Schölkopf et al., 999), kernel clustering (Ben-Hur et al., 2) or kernel Fisher discriminants (Mika et al., 999). To illustrate the use of kernel methods to analyze phylogenetic profiles, we test the ability of a Support Vector Machine to predict the functional class of a gene from its phylogenetic profile, and show that incorporating knowledge about the phylogenetic relationships between species into the kernel consistently improves its accuracy. In order to get a better insight of the relative positions of the phylogenetic profiles in the feature space, we also perform a kernel principal component analysis that shows that the tree kernel is more sensitive to evolutionary patterns than a kernel that does not incorporate any information about the phylogeny. METHODS AND ALGORITHMS Bayesian tree models for phylogenetic profiles The transmission of genes during evolution is modeled by a simple Bayesian tree model, i.e., a graphical model (Lauritzen, 996) based on a rooted tree. A rooted tree is a connected acyclic directed graph in which each node is l(u) c(u) u p(u) Fig.. A tree and the notations used in this article connected by a unique path from a designated node called the root and denoted. We denote by the set of nodes of the tree, the set of leaves (i.e., the nodes with no child), and the set of nodes without the root. For any node we write to represent the parent node of, and for any node, and to represent respectively the set of children of " # the node, and the set of leaves descendants of (Figure ). In this paper the tree is meant to be a phylogenetic tree: each leaf of the tree represents one organism currently living, and each internal node represents an ancestor of the current organisms (see for example Figure 3). The tree is used to define a joint probability distribution for a set of random variables %'&()' indexed by the nodes of the tree and with values in a finite alphabet*. We will focus on binary variables, i.e., *,-(/., but this work could be generalized to any finite alphabet. The is supposed to be variable % & attached to a node whenever the corresponding organism has a homologue of the gene under study, otherwise. For convenience we will use the following notations in the sequel. For any set of nodes 2, %3 4 % & ()' 5 is the subset of variables indexed by, and673* 3 a particular set of values that %83 can take. For any two sets of nodes 9 4 and ;:< 4, and any values 63==* 3 and > 3@? A* 3?, the notation B6 3DC > 3@? represents the following probability: 6E3 C >3? GF)HJI λ LKM5(N% & 6 & C KM : : ()% &? > &? PO With these notations a phylogenetic profile can be defined as follows: DEFINITION. A phylogenetic profile is a set of bits assigned to the leaves of the tree, i.e., 6RQS* Q. To model the evolution of genomes along the branches of the evolutionary tree an initial distribution MT on * 2
3 z H m Q Tree Kernel for Phylogenetic Profiles is assigned to the root of the tree, and a conditional probability distribution & U CWV for BUL( V X *'Y is assigned to each node Z'. The tree together with the set of distributions L & (N' defines a joint probability distribution for % [ as follows: KR6\[]* [ (^6_[<` T 6 T &ba [Ec & B6 & C 6ed f &hg )O Kernels Our approach to analyze phylogenetic profiles is to map them to a high-dimensional space whose dimensions correspond to evolutionarily relevant features. Let us denote by ij * Qlk m such a mapping, where n is the dimension of the feature space. Then the image of a profile 6_Qop* Q can be written as i B6Q_A i5q B6EQ\)(/OrO/Os( i mtb6_qu, and each coordinate itv 6EQ for Uw x.(/oro/os( n is called a feature of the profile 6eQ. In the sequel we consider a large set of evolutionarily relevant features, leading to a high-dimensional feature space. In such a case, rather than computing explicitly the image i 6 Q of each profile 6 Q * Q, it is more interesting to be able to compute efficiently the inner product between the images of any two profiles B6 Q ()> Q P* Q y * Q in the feature space, i.e., 6 Q ()> Q FNH{I v} 7q i v B6 Q i v B> Q )O () z The resulting function uo~(ro is called a kernel (Schölkopf & Smola, 2) and can be used to perform implicitly various data analysis algorithms in the feature space. The simplest kernel for phylogenetic profiles is the inner product when strings of bits are considered as real-valued vectors. In other words the naive kernel is defined as follows: z r v ƒ 6 Q (N> Q < 6 & > & O (2) &balq The resulting squared Euclidean distance between two profiles is simply the number of organism where they differ, hence the naive kernel is the natural way to represent profiles geometrically when their similarity is defined in such a way. The tree kernel The naive kernel (2) does not incorporate any knowledge about the nature of phylogenetic profiles, in particular the phylogenetic relationships among species. In order to develop a more biologically relevant measure of similarity it would be natural not to compare just the profiles themselves, but also their global patterns of inheritance if Fig. 2. An evolution pattern is a subtree (represented with thick lines) whose nodes are assigned values in J ˆ Š. the genomes of all ancestors were available. In that case we could check, for any two genes, when in evolution they were transmitted together or not, and get a more pertinent view of how similar their phylogenetic histories are than by just comparing their phylogenetic profiles organism per organism. In order to create a distance for phylogenetic profiles that reflects the similarity between evolutions we propose to map any profile to a feature space where each feature corresponds to a particular pattern of evolution defined as follows: DEFINITION 2. An evolution pattern is a pair B (/Œr3\ where # is a subtree of the complete Bayesian tree (i.e., a connected subgraph which contains the root), and Œh3* 3 is a set of values attached to the nodes of (see an example on Figure 2) For each evolution pattern (rœ 3 we define a feature i 3Ž j * Q2k which ideally would be. for profiles whose histories match the pattern, and - otherwise. The history of the each profile (i.e., the assignment of a - or a. to each ancestor organism in the phylogenetic tree) is of course not known, but the probabilistic model of evolution described previously can help approximate these ideal features. One way to proceed is to replace the ideal -. assignment by the probability that the profile 6eQ be observed conditionally to the pattern Œ 3. We therefore define the feature i 3\Ž as follows: KM6 Q * ( i 3\Ž 6 Q GFNH{I B6 Q C Œh3uO (3) Of course there is a priori no reason to focus on only one particular subtree and one particular pattern Œ 3. To the contrary it is more interesting to look at as many 3
4 J.-P.Vert z O features as possible in order to characterize each profile. we If we denote by the set of all subtrees of therefore define the set of features as the features i 3\Ž corresponding to all possible subtrees D and patterns Œh3S* 3. Combining () and (3) we see that the resulting kernel is the following, for any two profiles 6 Q ()> Q * Q y * Q : 6 Q (N> Q P 3 al f [ g al 6 Q C ŒL3B> Q C Œh3\)O In this expression all features are given the same importance in the sense that the kernel is obtained by adding the contributions of all features. However some features might be considered more important than others, because some evolution patterns are more likely to have effectively occurred than others. In other words it is likely that most features corresponding to patterns B (/Œ 3\ which have a very low probability of appearance are irrelevant and only add noise to the feature space. One solution to reduce this effect is to weight the contribution of each feature in the kernel by the probability of the corresponding pattern, which results in the following kernel: DEFINITION 3. The tree kernel is the kernel defined for any phylogenetic profiles B6 Q (N> Q P* 3 y * 3 by: z ~š B6 Q (N> Q FNH{I HJH )ŒL3_B6 Q C Œh3BB> Q C Œh3\)O (4) 3 al f [ g ah In this form the tree kernel appears to be related to convolution kernels introduced by (Watkins, 999) and (Haussler, 999), and to generalize the class of kernels developed in (Vert, 22) to graphical models with latent variables. Computation of the tree kernel Whereas the naive kernel between two profiles can easily be computed from (2), Equation (4) should not be directly implemented to compute the tree kernel because of the large number of terms to add (the number of subtrees of a tree increases exponentially with the number of nodes, and for each subtree there are sœ3 œ features to compute). In this section we present an algorithm to compute the tree kernel with a complexity linear with respect to the size of the phylogenetic profiles. Given a Bayesian tree model, the algorithm first computes the following functions, indexed by the nodes of the tree, for each phylogenetic profile 6 Q using a postorder traversal of the tree: If is a leaf: KMUw *( v 6 Q ()7 & B6 & C UL)O If is an internal node different from the root: KMUw*( sv 6EQ_()7 s& V C Uh 6EQ_() : uo ah &? a f &hg If x : B6_Q\(/ 7 st V B6EQ\(N : )O al &? a f Thg In a second step, in order to compute the kernel between any two profiles 6 Q and > Q, a second functional indexed by the nodes of the tree is computed using a post-order traversal of the tree: If is a leaf: v ) P If is not a leaf: v P &? a f &hg al. if 6 & > & U, - otherwise. &? V"C UL B : v 6 Q () : v > Q () : Finally the kernel value is obtained as follows: z ~š B6 Q (N6 Q P HJH al T V ) \ 6 Q (/ \ B> Q (r \)O This algorithm is based on post-order traversals of the tree and has therefore a complexity linear with respect to the size of the tree (and of the phylogenetic profiles). The reason why it returns the correct kernel value is a consequence of the following proposition which can be proved by induction by working up the tree from the leaves (due to space limitation the complete proof is omitted in this paper): PROPOSITION. The functions and computed by the previous algorithm satisfy the following equations, for any two phylogenetic profiles 6 Q ()> Q <* Q y * Q : For any node ' and Ut* : v 6 Q () B%S f &hg 6_ f &hg C %d f &hg UL)( and for the root: 6 Q (/ \G B% Q 6 Q )O For any node and Uw* : v P B%3 xœu3 C % & UL 3 ahª f &hg «al y B6e f &hg C Œh3\)O B>R f &hg C Œh3\)( where is the set of non-empty subtrees rooted in. 4
5 z Tree Kernel for Phylogenetic Profiles Support Vector Machines Support Vector Machines (SVMs) are a class of supervised learning algorithms first introduced by Vapnik and coworkers (Boser et al., 992; Vapnik, 998). Given a kernel z uo~(/o~ and a set of training examples (phylogenetic profiles in our case) labeled as positive or negative, SVMs learn a linear decision boundary in the feature space defined by the kernel in order to discriminate between positive and negative examples. Any new unlabeled example is then predicted to be positive or negative depending on the position of its image in the feature space relatively to the linear boundary. Apart from having strong mathematical foundations in statistical learning theory SVMs have been shown to perform very well in many real-world applications. They have found recently several applications in computational biology, including but not limited to protein function prediction (Jaakkola et al., 2), genes or tissues classification from microarray data (Brown et al., 2; Furey et al., 2) or translation initiation site recognition in DNA (Zien et al., 2). More details about the theory and practice of SVMs can be found in several books (Cristianini & Shawe-Taylor, 2; Schölkopf & Smola, 2). In this paper we use SVMs to infer the function of a gene from its phylogenetic profile. We test the ability of the tree kernel to incorporate biologically relevant information by comparing the performances of SVMs using either the tree kernel (4) or the naive kernel (2). We used the software SVMLight v.3.5 (Joachims, 999), a public implementation of SVMs that offers the possibility to implement user-defined kernels. Once a kernel is fixed only two parameters have to be specified before running the algorithm, namely the constant C which penalizes more or less misclassification errors, and the relative weights of positive and negative examples. To ensure a comparison as fair as possible between the naive and the tree kernels we used in all experiments the default value proposed by the software for the C parameter (namely Z v} q 6 f v g ()6 f v g for a training set 6 f q g (/OrO/Os(N6 f g ), and the relative weight parameter was adjusted to ensure that the total weight of positive examples be equal to the total weight of negative examples. Kernel PCA In order to get a glimpse of the relative positions of the phylogenetic profiles in the feature space defined by a kernel, it is possible to perform a principal component analysis (PCA) of their images in the feature space, and to look at their projections on the first few principal components. The PCA in the feature space, which is called a kernel PCA, can be performed entirely without computing the images of the profiles, but instead by computing the kernel between all pairs of profiles and then performing an eigenvalue decomposition of the resulting Gram matrix centered in the feature space (see details in (Schölkopf et al., 999)). We computed the Gram matrix with the C implementation of the kernel used by SVMLight, and performed the kernel-pca using the free and public mathematical software Octave±. DATA We performed experiments with the genes of the budding yeast Saccharomyces cerevisiae. Using the same data as (Pavlidis et al., 2) the phylogenetic profiles of 2465 yeast genes selected for their accurate functional classifications were generated by computing the E-value reported by BLAST version 2. (Altschul et al., 997) in a search against each of the 24 genomes listed in Figure 3 and collected from The Institute for Genomic Research website² or from the Sanger Center website³. Each bit in the phylogenetic tree was set to or if the E-value for the corresponding organism was larger or smaller than respectively. The phylogenetic tree used to compute the tree kernel, shown in Figure 3, is similar to the one used in (Liberles et al., 22). The parameters of the Bayesian model were arbitrarily set as follows: st). -O~ and for every node, R&u. C.µ &@u- C -t -O~. In other words this probabilistic model favors the transmission of genes along evolution but enables the appearance or disappearance of a gene along any branch with probability -O~.. In spite of its crudeness this simplistic model of gene transmission is sufficient to capture information about the phylogenetic relationships among different organisms. The function prediction experiments were based on the functional categories in the Munich Information Center for Protein Sequences Comprehensive Yeast Genome Databases (CYGD), whose functional catalog contains several hundreds of functional classes. We extracted all classes with at least genes, resulting in 33 classes. RESULTS Function prediction For each functional category we trained a SVM to predict whether a gene should be assigned to it or not based on its phylogenetic profile only. Using 3-fold cross validation repeated 5 times for each class, we compared the performances of a SVM using the naive kernel with a SVM using the tree kernel through there receiver operating characteristic (ROC) curves, i.e., the plot of true positives ± ² ³ 5
6 J.-P.Vert A. aeolicus T maritima D radiodurans Synechocystis B burgdorferi T pallidum C trachomatis C pneumoniae M pneumoniae M genitalium B subtilis M tuberculosis E coli H influenzae R prowazekii H pylori J99 H pylori A fulgidus M jannaschii M thermoautotrophicum P abyssi P horikoshii A pernix C elegans Fig. 3. The phylogenetic tree for 24 completely sequenced organisms (the lengths of the branches are arbitrary). as a function of false positives. When predicting the category of a gene, a SVM outputs a score (related to the distance between the example and the linear boundary in the feature space) so we could compute the ROC curves by varying a threshold and classifying the profiles depending on their scores compared to the threshold. As each functional class contains a small number of genes each learning problem is very unbalanced (there are few positive examples but many negative ones). We handled this issue by giving more weight to the positive examples in the SVM learning (such that the total weight of positive examples be equal to the total weight of negative examples). Moreover this implies that only a small percentage of false positives can be tolerated in realworld applications (such as function predictions), so we measured the ¹"º» ¼«½ score for each SVM, i.e., the area under the ROC curve up to the first 5 false positives (Gribskov & Robinson, 996). Table shows the functional categories with the highest ¹"º <¼«½ scores obtained by a SVM using the naive kernel, together with the ¹5º» ¼«½ scores of the SVM using the tree kernel. It turns out that the tree kernel performs better than the naive kernel on most of the classes, and sometimes improves considerably the ¹"º» <¼«½ score (for such classes as Glycolysis and gluconeogenesis, or trna modification). Classes that are better predicted by the naive kernel (amino-acid metabolism, transport facilitation, organization of plasma membrane and metabolism) tend to be larger and more general classes than the others. We plotted on Figures 4 the ROC curves up to 5 false positives corresponding to the four classes with the highest naive ¹"º ¼«½ score, in order to further study the differences in performance. These plots show that in several cases the tree kernel significantly outperforms the naive kernel when a very small number of false positives is allowed (for example, if ¾ false positives are accepted, the tree kernel retrieves on average more than 65% of all amino-acid transporters, while the naive kernel retrieves less than 3%). Kernel PCA analysis of four functional families In order to better understand the difference between the tree kernel and the naive kernel we performed a kernel PCA analysis of the phylogenetic profiles belonging to the first four categories listed in Table. Figure 5 shows the projections of these profiles on the first four principal components (PC) in the feature space defined by the tree and the naive kernel respectively. These plots show a clear difference between the geometries generated by both kernels. In the feature space defined by the naive kernel, the profiles are more scattered in all dimensions that in the feature space defined by the tree kernel. With the naive kernel, the profiles seem to form a convex set, but with the tree kernel, each PC seems to characterize particularly well a few profiles, which happen to belong to the same functional category (amino-acid transporters for the first component, or ABC transporters for the third one). The resulting shape looks like a small cluster at the center with many branches starting from the center toward various orthogonal directions, each direction being particularly relevant to one functional class. These observations show that the tree kernel is particularly sensitive to biologically relevant variations in the profiles. As an example, the ABC transporters (À ) characterized by a small value on the third PC with the tree kernel are also clustered together by the naive kernel (they have for instance a small value on the 4-th PC). However they are aligned along one particular PC with the tree kernel, which indicates that the variations among them are very typical of a neat combination of evolution patterns detected by the tree kernel. To the contrary their variations are spread among several PCs by the naive kernel. 6
7 Tree Kernel for Phylogenetic Profiles Table. ÁÂeÃeÄÆÅ scores for the prediction of 6 functional categories by a SVM using either a naive kernel or a tree kernel (the last column expresses the relative increase or decrease when the naive kernel is replaced by the tree kernel). Functional class Naive kernel Tree kernel Difference Amino-acid transporters % Fermentation % ABC transporters % C-compound, carbohydrate transport % Amino-acid biosynthesis % Amino-acid metabolism % Tricarboxylic-acid pathway % Transport Facilitation % Organization of plasma membrane.3.3-3% Amino-acid degradation (catabolism) % Lipid and fatty-acid transport % Homeostasis of other cations % Glycolysis and gluconeogenesis % Metabolism % Cellular import % trna modification % DISCUSSION AND CONCLUSION This paper presents an attempt to define a measure of similarity between phylogenetic profiles that incorporates knowledge about the phylogenetic relationships between species. More than just a measure of similarity, we were able to build a whole Euclidean geometry for phylogenetic profiles which is tractable in spite of its large dimension thanks to the availability of an efficient algorithm to compute the inner product in that space. Defining such a Euclidean geometry is more appealing than just defining a measure of similarity, because many data mining methods can be readily applied in such spaces. We demonstrated through a series of function prediction experiments and kernel PCA analysis that the geometry defined by the tree kernel is more sensitive to biologically relevant patterns in the profiles than the geometry generated when no evolutionary information is used. As a result a SVM using the tree kernel performed better on average than a SVM using a naive kernel. Because of the efficiency of kernel methods such as SVMs for classification, developing kernels for particular objects has received much attention recently. As an example, a number of kernels for strings have been developed and applied to the classification of biological sequences or text written in natural language, including the Fisher kernel (Jaakkola et al., 2), convolution kernels (Haussler, 999; Watkins, 999), string kernels (Lodhi et al., 2), interpolated kernels (Vert, 22) or spectrum kernels (Leslie et al., 22). More than a string kernel, the tree kernel introduced in this paper should be regarded as a graph kernel, as it is computed for sets of strings indexed by nodes of a tree.the algorithm can easily be generalized to handle any tree graphical model with or without latent variables, in order to define a kernel on the set of realizations of these graphical models. As such models are widely used not only in computational biology but also in many other fields, the potential applications of this kernel in combination with the increasing library of kernel methods for data mining are numerous. ACKNOWLEDGEMENTS I thank Minoru Kanehisa for helpful discussions and for supporting this research through a Research for the Future Program of the Japan Society for the Promotion of Science. REFERENCES Altschul, S., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. (997). Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Research, 25, Ben-Hur, A., Horn, D., Siegelmann, H. T. & Vapnik, V. (2). Support vector clustering. Journal of Machine Learning Research, 2, Boser, B. E., Guyon, I. M. & Vapnik, V. N. (992). A training algorithm for optimal margin classifiers. In Proceedings of the 5th annual ACM workshop on Computational Learning Theory. ACM Press, pp Brown, M. P. S., Grundy, W. N., Lin, D., Cristianini, N., Sugnet, C. W., Furey, T. S., Manuel Ares, J. & Haussler, D. (2). Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. USA, 97, Cristianini, N. & Shawe-Taylor, J. (2). An introduction 7
8 J.-P.Vert Amino acid transporters o : Tree : Naive Fermentation o : Tree : Naive ABC transporters o : Tree : Naive C compound, carbohydrate transport o : Tree : Naive Fig. 4. ROC curves for the prediction of four functional classes from the phylogenetic profiles of the yeast genes with a Support Vector Machine using the tree kernel (labeled tree ) or the naive kernel (labelled naive ). The number of false positives is on the X-axis, the percentage of true positives retrieved (i.e., the recall) on the Y-axis. Curves are averaged over a number of 3-fold cross-validation experiments. to Support Vector Machines and other kernel-based learning methods. Cambridge University Press. Furey, T. S., Duffy, N., Cristianini, N., Bednarski, D., Schummer, M. & Haussler, D. (2). Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics, 6, Gribskov, M. & Robinson, N. (996). Use of receiver operating characteristic (roc) analysis to evaluate sequence matching. Computers and Chemistry, 2, Haussler, D. (999). Convolution kernels on discrete structures. Technical report, UC Santa Crux. Jaakkola, T., Diekhans, M. & Haussler, D. (2). A discrimitive framework for detecting remote protein homologies. Journal of Computational Biology, 7, Joachims, T. (999). Making large-scale svm learning practical. In Schölkopf, B., Burges, C. & Smola, A., (eds.) Advances in Kernel Methods - Support Vector Learning. MIT Press, pp Lauritzen, S. (996). Graphical Models. Oxford. Leslie, C., Eskin, E. & Noble, W. S. (22). The spectrum kernel: a string kernel for svm protein classification. In Altman, R. B., Dunker, A. K., Hunter, L., Lauerdale, K. & Klein, T. E., (eds.) Proceedings of the Pacific Symposium on Biocomputing 22. World Scientific, pp Liberles, D. A., Thoren, A., von Heijne, G. & Elofsson, A. (22). The use of phylogenetic profiles for gene predictions. Current Genomics. In press. Lodhi, H., Shawe-Taylor, J., Cristianini, N. & Watkins, C. J. C. H. (2). Text classification using string kernels. In NIPS. pp Marcotte, E. M., Pellegrini, M., Thompson, M. J., Yeates, T. O. & Eisenberg, D. (999). A combined algorithm for genome-wide prediction of protein function. Nature, 42, Mika, S., Rätsch, G., Weston, J., Schölkopf, B. & Müller, K. (999). Fisher discriminant analysis with kernels. In Hu, Y.-H., Larsen, 8
9 Tree Kernel for Phylogenetic Profiles Tree kernel PCA (x=, y=2) Naive kernel PCA (x=, y=2) Tree kernel PCA (x=3, y=4) Naive kernel PCA (x=3, y=4) Fig. 5. Kernel PCA analysis of the phylogenetic profiles of the genes belonging to four functional classes, with either the tree kernel (on the left) or the naive kernel (on the right). In each case the upper plot represents the first PC versus the second, and the lower plot represents the third against the fourth. Each symbol represents one functional class: amino-acid transporters (Ç ), fermentation (È ), ABC transporters (É ) and C-compound, carbohydrate transport (Ê ) J., Wilson, E. & Douglas, S., (eds.) Neural Networks for Signal Processing IX. IEEE, pp Pavlidis, P., Weston, J., Cai, J. & Grundy, W. N. (2). Gene functional classification from heterogeneous data. In Proceedings of the Fifth Annual International Conference on Computational Biology. pp Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. (999). Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA, 96, Schölkopf, B. & Smola, A. J. (2). Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. MIT Press. Schölkopf, B., Smola, A. J. & Müller, K.-R. (999). Kernel principal component analysis. In Schölkopf, B., Burges, C. & Smola, A., (eds.) Advances in Kernel Methods - Support Vector Learning. MIT Press, pp Vapnik, V. N. (998). Statistical Learning Theory. Wiley, New- York. Vert, J.-P. (22). Support vector machine prediction of signal peptide cleavage site using a new class of kernels for strings. In Altman, R. B., Dunker, A. K., Hunter, L., Lauerdale, K. & Klein, T. E., (eds.) Proceedings of the Pacific Symposium on Biocomputing 22. World Scientific, pp Watkins, C. (999). Dynamic alignment kernels. Technical report, UL Royal Holloway. Zien, A., Ratsch, G., Mika, S., Scholkopf, B., Lengauer, T. & Muller, K.-R. (2). Engineering support vector machine kernels that recognize translation initiation sites. Bioinformatics, 6,
Transductive learning with EM algorithm to classify proteins based on phylogenetic profiles
 Int. J. Data Mining and Bioinformatics, Vol. 1, No. 4, 2007 337 Transductive learning with EM algorithm to classify proteins based on phylogenetic profiles Roger A. Craig and Li Liao* Department of Computer
Int. J. Data Mining and Bioinformatics, Vol. 1, No. 4, 2007 337 Transductive learning with EM algorithm to classify proteins based on phylogenetic profiles Roger A. Craig and Li Liao* Department of Computer
Support Vector Machines (SVM) in bioinformatics. Day 1: Introduction to SVM
 1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
The Minimal-Gene-Set -Kapil PHY498BIO, HW 3
 The Minimal-Gene-Set -Kapil Rajaraman(rajaramn@uiuc.edu) PHY498BIO, HW 3 The number of genes in organisms varies from around 480 (for parasitic bacterium Mycoplasma genitalium) to the order of 100,000
The Minimal-Gene-Set -Kapil Rajaraman(rajaramn@uiuc.edu) PHY498BIO, HW 3 The number of genes in organisms varies from around 480 (for parasitic bacterium Mycoplasma genitalium) to the order of 100,000
Support vector machines, Kernel methods, and Applications in bioinformatics
 1 Support vector machines, Kernel methods, and Applications in bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Machine Learning in Bioinformatics conference,
1 Support vector machines, Kernel methods, and Applications in bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Machine Learning in Bioinformatics conference,
Combining pairwise sequence similarity and support vector machines for remote protein homology detection
 Combining pairwise sequence similarity and support vector machines for remote protein homology detection Li Liao Central Research & Development E. I. du Pont de Nemours Company li.liao@usa.dupont.com William
Combining pairwise sequence similarity and support vector machines for remote protein homology detection Li Liao Central Research & Development E. I. du Pont de Nemours Company li.liao@usa.dupont.com William
2 Genome evolution: gene fusion versus gene fission
 2 Genome evolution: gene fusion versus gene fission Berend Snel, Peer Bork and Martijn A. Huynen Trends in Genetics 16 (2000) 9-11 13 Chapter 2 Introduction With the advent of complete genome sequencing,
2 Genome evolution: gene fusion versus gene fission Berend Snel, Peer Bork and Martijn A. Huynen Trends in Genetics 16 (2000) 9-11 13 Chapter 2 Introduction With the advent of complete genome sequencing,
Support Vector Machines.
 Support Vector Machines www.cs.wisc.edu/~dpage 1 Goals for the lecture you should understand the following concepts the margin slack variables the linear support vector machine nonlinear SVMs the kernel
Support Vector Machines www.cs.wisc.edu/~dpage 1 Goals for the lecture you should understand the following concepts the margin slack variables the linear support vector machine nonlinear SVMs the kernel
Combining pairwise sequence similarity and support vector machines for remote protein homology detection
 Combining pairwise sequence similarity and support vector machines for remote protein homology detection Li Liao Central Research & Development E. I. du Pont de Nemours Company li.liao@usa.dupont.com William
Combining pairwise sequence similarity and support vector machines for remote protein homology detection Li Liao Central Research & Development E. I. du Pont de Nemours Company li.liao@usa.dupont.com William
Kernels for gene regulatory regions
 Kernels for gene regulatory regions Jean-Philippe Vert Geostatistics Center Ecole des Mines de Paris - ParisTech Jean-Philippe.Vert@ensmp.fr Robert Thurman Division of Medical Genetics University of Washington
Kernels for gene regulatory regions Jean-Philippe Vert Geostatistics Center Ecole des Mines de Paris - ParisTech Jean-Philippe.Vert@ensmp.fr Robert Thurman Division of Medical Genetics University of Washington
Linear Classification and SVM. Dr. Xin Zhang
 Linear Classification and SVM Dr. Xin Zhang Email: eexinzhang@scut.edu.cn What is linear classification? Classification is intrinsically non-linear It puts non-identical things in the same class, so a
Linear Classification and SVM Dr. Xin Zhang Email: eexinzhang@scut.edu.cn What is linear classification? Classification is intrinsically non-linear It puts non-identical things in the same class, so a
Microarray Data Analysis: Discovery
 Microarray Data Analysis: Discovery Lecture 5 Classification Classification vs. Clustering Classification: Goal: Placing objects (e.g. genes) into meaningful classes Supervised Clustering: Goal: Discover
Microarray Data Analysis: Discovery Lecture 5 Classification Classification vs. Clustering Classification: Goal: Placing objects (e.g. genes) into meaningful classes Supervised Clustering: Goal: Discover
Biological Networks: Comparison, Conservation, and Evolution via Relative Description Length By: Tamir Tuller & Benny Chor
 Biological Networks:,, and via Relative Description Length By: Tamir Tuller & Benny Chor Presented by: Noga Grebla Content of the presentation Presenting the goals of the research Reviewing basic terms
Biological Networks:,, and via Relative Description Length By: Tamir Tuller & Benny Chor Presented by: Noga Grebla Content of the presentation Presenting the goals of the research Reviewing basic terms
A Study of Network-based Kernel Methods on Protein-Protein Interaction for Protein Functions Prediction
 The Third International Symposium on Optimization and Systems Biology (OSB 09) Zhangjiajie, China, September 20 22, 2009 Copyright 2009 ORSC & APORC, pp. 25 32 A Study of Network-based Kernel Methods on
The Third International Symposium on Optimization and Systems Biology (OSB 09) Zhangjiajie, China, September 20 22, 2009 Copyright 2009 ORSC & APORC, pp. 25 32 A Study of Network-based Kernel Methods on
Sequence Alignment Techniques and Their Uses
 Sequence Alignment Techniques and Their Uses Sarah Fiorentino Since rapid sequencing technology and whole genomes sequencing, the amount of sequence information has grown exponentially. With all of this
Sequence Alignment Techniques and Their Uses Sarah Fiorentino Since rapid sequencing technology and whole genomes sequencing, the amount of sequence information has grown exponentially. With all of this
Support'Vector'Machines. Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan
 Support'Vector'Machines Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan kasthuri.kannan@nyumc.org Overview Support Vector Machines for Classification Linear Discrimination Nonlinear Discrimination
Support'Vector'Machines Machine(Learning(Spring(2018 March(5(2018 Kasthuri Kannan kasthuri.kannan@nyumc.org Overview Support Vector Machines for Classification Linear Discrimination Nonlinear Discrimination
Support Vector Machine & Its Applications
 Support Vector Machine & Its Applications A portion (1/3) of the slides are taken from Prof. Andrew Moore s SVM tutorial at http://www.cs.cmu.edu/~awm/tutorials Mingyue Tan The University of British Columbia
Support Vector Machine & Its Applications A portion (1/3) of the slides are taken from Prof. Andrew Moore s SVM tutorial at http://www.cs.cmu.edu/~awm/tutorials Mingyue Tan The University of British Columbia
Kernel Methods and Support Vector Machines
 Kernel Methods and Support Vector Machines Bernhard Schölkopf Max-Planck-Institut für biologische Kybernetik 72076 Tübingen, Germany Bernhard.Schoelkopf@tuebingen.mpg.de Alex Smola RSISE, Australian National
Kernel Methods and Support Vector Machines Bernhard Schölkopf Max-Planck-Institut für biologische Kybernetik 72076 Tübingen, Germany Bernhard.Schoelkopf@tuebingen.mpg.de Alex Smola RSISE, Australian National
# shared OGs (spa, spb) Size of the smallest genome. dist (spa, spb) = 1. Neighbor joining. OG1 OG2 OG3 OG4 sp sp sp
 Bioinformatics and Evolutionary Genomics: Genome Evolution in terms of Gene Content 3/10/2014 1 Gene Content Evolution What about HGT / genome sizes? Genome trees based on gene content: shared genes Haemophilus
Bioinformatics and Evolutionary Genomics: Genome Evolution in terms of Gene Content 3/10/2014 1 Gene Content Evolution What about HGT / genome sizes? Genome trees based on gene content: shared genes Haemophilus
Discriminative Direction for Kernel Classifiers
 Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Genome Annotation. Bioinformatics and Computational Biology. Genome sequencing Assembly. Gene prediction. Protein targeting.
 Genome Annotation Bioinformatics and Computational Biology Genome Annotation Frank Oliver Glöckner 1 Genome Analysis Roadmap Genome sequencing Assembly Gene prediction Protein targeting trna prediction
Genome Annotation Bioinformatics and Computational Biology Genome Annotation Frank Oliver Glöckner 1 Genome Analysis Roadmap Genome sequencing Assembly Gene prediction Protein targeting trna prediction
Internet Electronic Journal of Molecular Design
 ISSN 1538 6414 Internet Electronic Journal of Molecular Design May 2003, Volume 2, Number 5, Pages 348 357 Editor: Ovidiu Ivanciuc Special issue dedicated to Professor Haruo Hosoya on the occasion of the
ISSN 1538 6414 Internet Electronic Journal of Molecular Design May 2003, Volume 2, Number 5, Pages 348 357 Editor: Ovidiu Ivanciuc Special issue dedicated to Professor Haruo Hosoya on the occasion of the
Introduction to Support Vector Machines
 Introduction to Support Vector Machines Andreas Maletti Technische Universität Dresden Fakultät Informatik June 15, 2006 1 The Problem 2 The Basics 3 The Proposed Solution Learning by Machines Learning
Introduction to Support Vector Machines Andreas Maletti Technische Universität Dresden Fakultät Informatik June 15, 2006 1 The Problem 2 The Basics 3 The Proposed Solution Learning by Machines Learning
Metabolic networks: Activity detection and Inference
 1 Metabolic networks: Activity detection and Inference Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Advanced microarray analysis course, Elsinore, Denmark, May 21th,
1 Metabolic networks: Activity detection and Inference Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group Advanced microarray analysis course, Elsinore, Denmark, May 21th,
Analysis of N-terminal Acetylation data with Kernel-Based Clustering
 Analysis of N-terminal Acetylation data with Kernel-Based Clustering Ying Liu Department of Computational Biology, School of Medicine University of Pittsburgh yil43@pitt.edu 1 Introduction N-terminal acetylation
Analysis of N-terminal Acetylation data with Kernel-Based Clustering Ying Liu Department of Computational Biology, School of Medicine University of Pittsburgh yil43@pitt.edu 1 Introduction N-terminal acetylation
Computational approaches for functional genomics
 Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie
 A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
Modifying A Linear Support Vector Machine for Microarray Data Classification
 Modifying A Linear Support Vector Machine for Microarray Data Classification Prof. Rosemary A. Renaut Dr. Hongbin Guo & Wang Juh Chen Department of Mathematics and Statistics, Arizona State University
Modifying A Linear Support Vector Machine for Microarray Data Classification Prof. Rosemary A. Renaut Dr. Hongbin Guo & Wang Juh Chen Department of Mathematics and Statistics, Arizona State University
Bayesian Data Fusion with Gaussian Process Priors : An Application to Protein Fold Recognition
 Bayesian Data Fusion with Gaussian Process Priors : An Application to Protein Fold Recognition Mar Girolami 1 Department of Computing Science University of Glasgow girolami@dcs.gla.ac.u 1 Introduction
Bayesian Data Fusion with Gaussian Process Priors : An Application to Protein Fold Recognition Mar Girolami 1 Department of Computing Science University of Glasgow girolami@dcs.gla.ac.u 1 Introduction
SUPPORT VECTOR REGRESSION WITH A GENERALIZED QUADRATIC LOSS
 SUPPORT VECTOR REGRESSION WITH A GENERALIZED QUADRATIC LOSS Filippo Portera and Alessandro Sperduti Dipartimento di Matematica Pura ed Applicata Universit a di Padova, Padova, Italy {portera,sperduti}@math.unipd.it
SUPPORT VECTOR REGRESSION WITH A GENERALIZED QUADRATIC LOSS Filippo Portera and Alessandro Sperduti Dipartimento di Matematica Pura ed Applicata Universit a di Padova, Padova, Italy {portera,sperduti}@math.unipd.it
Computational methods for predicting protein-protein interactions
 Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference
 CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
Carson Andorf 1,3, Adrian Silvescu 1,3, Drena Dobbs 2,3,4, Vasant Honavar 1,3,4. University, Ames, Iowa, 50010, USA. Ames, Iowa, 50010, USA
 Learning Classifiers for Assigning Protein Sequences to Gene Ontology Functional Families: Combining of Function Annotation Using Sequence Homology With that Based on Amino Acid k-gram Composition Yields
Learning Classifiers for Assigning Protein Sequences to Gene Ontology Functional Families: Combining of Function Annotation Using Sequence Homology With that Based on Amino Acid k-gram Composition Yields
On learning with kernels for unordered pairs
 Martial Hue MartialHue@mines-paristechfr Jean-Philippe Vert Jean-PhilippeVert@mines-paristechfr Mines ParisTech, Centre for Computational Biology, 35 rue Saint Honoré, F-77300 Fontainebleau, France Institut
Martial Hue MartialHue@mines-paristechfr Jean-Philippe Vert Jean-PhilippeVert@mines-paristechfr Mines ParisTech, Centre for Computational Biology, 35 rue Saint Honoré, F-77300 Fontainebleau, France Institut
Analysis of Multiclass Support Vector Machines
 Analysis of Multiclass Support Vector Machines Shigeo Abe Graduate School of Science and Technology Kobe University Kobe, Japan abe@eedept.kobe-u.ac.jp Abstract Since support vector machines for pattern
Analysis of Multiclass Support Vector Machines Shigeo Abe Graduate School of Science and Technology Kobe University Kobe, Japan abe@eedept.kobe-u.ac.jp Abstract Since support vector machines for pattern
CISC 636 Computational Biology & Bioinformatics (Fall 2016)
 CISC 636 Computational Biology & Bioinformatics (Fall 2016) Predicting Protein-Protein Interactions CISC636, F16, Lec22, Liao 1 Background Proteins do not function as isolated entities. Protein-Protein
CISC 636 Computational Biology & Bioinformatics (Fall 2016) Predicting Protein-Protein Interactions CISC636, F16, Lec22, Liao 1 Background Proteins do not function as isolated entities. Protein-Protein
Kernel expansions with unlabeled examples
 Kernel expansions with unlabeled examples Martin Szummer MIT AI Lab & CBCL Cambridge, MA szummer@ai.mit.edu Tommi Jaakkola MIT AI Lab Cambridge, MA tommi@ai.mit.edu Abstract Modern classification applications
Kernel expansions with unlabeled examples Martin Szummer MIT AI Lab & CBCL Cambridge, MA szummer@ai.mit.edu Tommi Jaakkola MIT AI Lab Cambridge, MA tommi@ai.mit.edu Abstract Modern classification applications
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
species, if their corresponding mrnas share similar expression patterns, or if the proteins interact with one another. It seems natural that, while al
 KERNEL-BASED DATA FUSION AND ITS APPLICATION TO PROTEIN FUNCTION PREDICTION IN YEAST GERT R. G. LANCKRIET Division of Electrical Engineering, University of California, Berkeley MINGHUA DENG Department
KERNEL-BASED DATA FUSION AND ITS APPLICATION TO PROTEIN FUNCTION PREDICTION IN YEAST GERT R. G. LANCKRIET Division of Electrical Engineering, University of California, Berkeley MINGHUA DENG Department
DIPARTIMENTO DI INGEGNERIA E SCIENZA DELL INFORMAZIONE Povo Trento (Italy), Via Sommarive 14
 UNIVERSITY OF TRENTO DIPARTIMENTO DI INGEGNERIA E SCIENZA DELL INFORMAZIONE 38050 Povo Trento (Italy), Via Sommarive 14 http://www.disi.unitn.it KERNEL INTEGRATION USING VON NEUMANN ENTROPY Andrea Malossini,
UNIVERSITY OF TRENTO DIPARTIMENTO DI INGEGNERIA E SCIENZA DELL INFORMAZIONE 38050 Povo Trento (Italy), Via Sommarive 14 http://www.disi.unitn.it KERNEL INTEGRATION USING VON NEUMANN ENTROPY Andrea Malossini,
SVM-Fold: a tool for discriminative multi-class protein fold and superfamily recognition
 SVM-Fold: a tool for discriminative multi-class protein fold and superfamily recognition Iain Melvin 1,2, Eugene Ie 3, Rui Kuang 4, Jason Weston 1, William Stafford Noble 5, Christina Leslie 2,6 1 NEC
SVM-Fold: a tool for discriminative multi-class protein fold and superfamily recognition Iain Melvin 1,2, Eugene Ie 3, Rui Kuang 4, Jason Weston 1, William Stafford Noble 5, Christina Leslie 2,6 1 NEC
Cluster Kernels for Semi-Supervised Learning
 Cluster Kernels for Semi-Supervised Learning Olivier Chapelle, Jason Weston, Bernhard Scholkopf Max Planck Institute for Biological Cybernetics, 72076 Tiibingen, Germany {first. last} @tuebingen.mpg.de
Cluster Kernels for Semi-Supervised Learning Olivier Chapelle, Jason Weston, Bernhard Scholkopf Max Planck Institute for Biological Cybernetics, 72076 Tiibingen, Germany {first. last} @tuebingen.mpg.de
Support Vector Machines
 Support Vector Machines Reading: Ben-Hur & Weston, A User s Guide to Support Vector Machines (linked from class web page) Notation Assume a binary classification problem. Instances are represented by vector
Support Vector Machines Reading: Ben-Hur & Weston, A User s Guide to Support Vector Machines (linked from class web page) Notation Assume a binary classification problem. Instances are represented by vector
2 GENE FUNCTIONAL SIMILARITY. 2.1 Semantic values of GO terms
 Bioinformatics Advance Access published March 7, 2007 The Author (2007). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Bioinformatics Advance Access published March 7, 2007 The Author (2007). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Manifold Regularization
 Manifold Regularization Vikas Sindhwani Department of Computer Science University of Chicago Joint Work with Mikhail Belkin and Partha Niyogi TTI-C Talk September 14, 24 p.1 The Problem of Learning is
Manifold Regularization Vikas Sindhwani Department of Computer Science University of Chicago Joint Work with Mikhail Belkin and Partha Niyogi TTI-C Talk September 14, 24 p.1 The Problem of Learning is
Introduction to Bioinformatics Integrated Science, 11/9/05
 1 Introduction to Bioinformatics Integrated Science, 11/9/05 Morris Levy Biological Sciences Research: Evolutionary Ecology, Plant- Fungal Pathogen Interactions Coordinator: BIOL 495S/CS490B/STAT490B Introduction
1 Introduction to Bioinformatics Integrated Science, 11/9/05 Morris Levy Biological Sciences Research: Evolutionary Ecology, Plant- Fungal Pathogen Interactions Coordinator: BIOL 495S/CS490B/STAT490B Introduction
Multiple Similarities Based Kernel Subspace Learning for Image Classification
 Multiple Similarities Based Kernel Subspace Learning for Image Classification Wang Yan, Qingshan Liu, Hanqing Lu, and Songde Ma National Laboratory of Pattern Recognition, Institute of Automation, Chinese
Multiple Similarities Based Kernel Subspace Learning for Image Classification Wang Yan, Qingshan Liu, Hanqing Lu, and Songde Ma National Laboratory of Pattern Recognition, Institute of Automation, Chinese
SUPERVISED LEARNING: INTRODUCTION TO CLASSIFICATION
 SUPERVISED LEARNING: INTRODUCTION TO CLASSIFICATION 1 Outline Basic terminology Features Training and validation Model selection Error and loss measures Statistical comparison Evaluation measures 2 Terminology
SUPERVISED LEARNING: INTRODUCTION TO CLASSIFICATION 1 Outline Basic terminology Features Training and validation Model selection Error and loss measures Statistical comparison Evaluation measures 2 Terminology
CHAPTER-17. Decision Tree Induction
 CHAPTER-17 Decision Tree Induction 17.1 Introduction 17.2 Attribute selection measure 17.3 Tree Pruning 17.4 Extracting Classification Rules from Decision Trees 17.5 Bayesian Classification 17.6 Bayes
CHAPTER-17 Decision Tree Induction 17.1 Introduction 17.2 Attribute selection measure 17.3 Tree Pruning 17.4 Extracting Classification Rules from Decision Trees 17.5 Bayesian Classification 17.6 Bayes
Machine learning for pervasive systems Classification in high-dimensional spaces
 Machine learning for pervasive systems Classification in high-dimensional spaces Department of Communications and Networking Aalto University, School of Electrical Engineering stephan.sigg@aalto.fi Version
Machine learning for pervasive systems Classification in high-dimensional spaces Department of Communications and Networking Aalto University, School of Electrical Engineering stephan.sigg@aalto.fi Version
Representing structured relational data in Euclidean vector spaces. Tony Plate
 Representing structured relational data in Euclidean vector spaces Tony Plate tplate@acm.org http://www.d-reps.org October 2004 AAAI Symposium 2004 1 Overview A general method for representing structured
Representing structured relational data in Euclidean vector spaces Tony Plate tplate@acm.org http://www.d-reps.org October 2004 AAAI Symposium 2004 1 Overview A general method for representing structured
Introduction to Bioinformatics
 CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
Linking non-binned spike train kernels to several existing spike train metrics
 Linking non-binned spike train kernels to several existing spike train metrics Benjamin Schrauwen Jan Van Campenhout ELIS, Ghent University, Belgium Benjamin.Schrauwen@UGent.be Abstract. This work presents
Linking non-binned spike train kernels to several existing spike train metrics Benjamin Schrauwen Jan Van Campenhout ELIS, Ghent University, Belgium Benjamin.Schrauwen@UGent.be Abstract. This work presents
The Homology Kernel: A Biologically Motivated Sequence Embedding into Euclidean Space
 The Homology Kernel: A Biologically Motivated Sequence Embedding into Euclidean Space Eleazar Eskin Department of Computer Science Engineering University of California, San Diego eeskin@cs.ucsd.edu Sagi
The Homology Kernel: A Biologically Motivated Sequence Embedding into Euclidean Space Eleazar Eskin Department of Computer Science Engineering University of California, San Diego eeskin@cs.ucsd.edu Sagi
below, kernel PCA Eigenvectors, and linear combinations thereof. For the cases where the pre-image does exist, we can provide a means of constructing
 Kernel PCA Pattern Reconstruction via Approximate Pre-Images Bernhard Scholkopf, Sebastian Mika, Alex Smola, Gunnar Ratsch, & Klaus-Robert Muller GMD FIRST, Rudower Chaussee 5, 12489 Berlin, Germany fbs,
Kernel PCA Pattern Reconstruction via Approximate Pre-Images Bernhard Scholkopf, Sebastian Mika, Alex Smola, Gunnar Ratsch, & Klaus-Robert Muller GMD FIRST, Rudower Chaussee 5, 12489 Berlin, Germany fbs,
K-means-based Feature Learning for Protein Sequence Classification
 K-means-based Feature Learning for Protein Sequence Classification Paul Melman and Usman W. Roshan Department of Computer Science, NJIT Newark, NJ, 07102, USA pm462@njit.edu, usman.w.roshan@njit.edu Abstract
K-means-based Feature Learning for Protein Sequence Classification Paul Melman and Usman W. Roshan Department of Computer Science, NJIT Newark, NJ, 07102, USA pm462@njit.edu, usman.w.roshan@njit.edu Abstract
BMD645. Integration of Omics
 BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
10-810: Advanced Algorithms and Models for Computational Biology. Optimal leaf ordering and classification
 10-810: Advanced Algorithms and Models for Computational Biology Optimal leaf ordering and classification Hierarchical clustering As we mentioned, its one of the most popular methods for clustering gene
10-810: Advanced Algorithms and Models for Computational Biology Optimal leaf ordering and classification Hierarchical clustering As we mentioned, its one of the most popular methods for clustering gene
Learning the Semantic Correlation: An Alternative Way to Gain from Unlabeled Text
 Learning the Semantic Correlation: An Alternative Way to Gain from Unlabeled Text Yi Zhang Machine Learning Department Carnegie Mellon University yizhang1@cs.cmu.edu Jeff Schneider The Robotics Institute
Learning the Semantic Correlation: An Alternative Way to Gain from Unlabeled Text Yi Zhang Machine Learning Department Carnegie Mellon University yizhang1@cs.cmu.edu Jeff Schneider The Robotics Institute
Formulation with slack variables
 Formulation with slack variables Optimal margin classifier with slack variables and kernel functions described by Support Vector Machine (SVM). min (w,ξ) ½ w 2 + γσξ(i) subject to ξ(i) 0 i, d(i) (w T x(i)
Formulation with slack variables Optimal margin classifier with slack variables and kernel functions described by Support Vector Machine (SVM). min (w,ξ) ½ w 2 + γσξ(i) subject to ξ(i) 0 i, d(i) (w T x(i)
SVM TRADE-OFF BETWEEN MAXIMIZE THE MARGIN AND MINIMIZE THE VARIABLES USED FOR REGRESSION
 International Journal of Pure and Applied Mathematics Volume 87 No. 6 2013, 741-750 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu doi: http://dx.doi.org/10.12732/ijpam.v87i6.2
International Journal of Pure and Applied Mathematics Volume 87 No. 6 2013, 741-750 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu doi: http://dx.doi.org/10.12732/ijpam.v87i6.2
Prediction of the Risk Types of Human Papillomaviruses by Support Vector Machines
 Prediction of the Risk Types of Human Papillomaviruses by Support Vector Machines Je-Gun Joung 1,2, Sok June O 2,4, and Byoung-Tak Zhang 1,2,3 1 Biointelligence Laboratory, Graduate Program in Bioinformatics
Prediction of the Risk Types of Human Papillomaviruses by Support Vector Machines Je-Gun Joung 1,2, Sok June O 2,4, and Byoung-Tak Zhang 1,2,3 1 Biointelligence Laboratory, Graduate Program in Bioinformatics
Motif Extraction and Protein Classification
 Motif Extraction and Protein Classification Vered Kunik 1 Zach Solan 2 Shimon Edelman 3 Eytan Ruppin 1 David Horn 2 1 School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel {kunikver,ruppin}@tau.ac.il
Motif Extraction and Protein Classification Vered Kunik 1 Zach Solan 2 Shimon Edelman 3 Eytan Ruppin 1 David Horn 2 1 School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel {kunikver,ruppin}@tau.ac.il
Computational Biology: Basics & Interesting Problems
 Computational Biology: Basics & Interesting Problems Summary Sources of information Biological concepts: structure & terminology Sequencing Gene finding Protein structure prediction Sources of information
Computational Biology: Basics & Interesting Problems Summary Sources of information Biological concepts: structure & terminology Sequencing Gene finding Protein structure prediction Sources of information
December 2, :4 WSPC/INSTRUCTION FILE jbcb-profile-kernel. Profile-based string kernels for remote homology detection and motif extraction
 Journal of Bioinformatics and Computational Biology c Imperial College Press Profile-based string kernels for remote homology detection and motif extraction Rui Kuang 1, Eugene Ie 1,3, Ke Wang 1, Kai Wang
Journal of Bioinformatics and Computational Biology c Imperial College Press Profile-based string kernels for remote homology detection and motif extraction Rui Kuang 1, Eugene Ie 1,3, Ke Wang 1, Kai Wang
Scale-Invariance of Support Vector Machines based on the Triangular Kernel. Abstract
 Scale-Invariance of Support Vector Machines based on the Triangular Kernel François Fleuret Hichem Sahbi IMEDIA Research Group INRIA Domaine de Voluceau 78150 Le Chesnay, France Abstract This paper focuses
Scale-Invariance of Support Vector Machines based on the Triangular Kernel François Fleuret Hichem Sahbi IMEDIA Research Group INRIA Domaine de Voluceau 78150 Le Chesnay, France Abstract This paper focuses
Universal Learning Technology: Support Vector Machines
 Special Issue on Information Utilizing Technologies for Value Creation Universal Learning Technology: Support Vector Machines By Vladimir VAPNIK* This paper describes the Support Vector Machine (SVM) technology,
Special Issue on Information Utilizing Technologies for Value Creation Universal Learning Technology: Support Vector Machines By Vladimir VAPNIK* This paper describes the Support Vector Machine (SVM) technology,
Efficient Remote Homology Detection with Secondary Structure
 Efficient Remote Homology Detection with Secondary Structure 2 Yuna Hou 1, Wynne Hsu 1, Mong Li Lee 1, and Christopher Bystroff 2 1 School of Computing,National University of Singapore,Singapore 117543
Efficient Remote Homology Detection with Secondary Structure 2 Yuna Hou 1, Wynne Hsu 1, Mong Li Lee 1, and Christopher Bystroff 2 1 School of Computing,National University of Singapore,Singapore 117543
On the Sizes of Decision Diagrams Representing the Set of All Parse Trees of a Context-free Grammar
 Proceedings of Machine Learning Research vol 73:153-164, 2017 AMBN 2017 On the Sizes of Decision Diagrams Representing the Set of All Parse Trees of a Context-free Grammar Kei Amii Kyoto University Kyoto
Proceedings of Machine Learning Research vol 73:153-164, 2017 AMBN 2017 On the Sizes of Decision Diagrams Representing the Set of All Parse Trees of a Context-free Grammar Kei Amii Kyoto University Kyoto
Support Vector Machines
 Support Vector Machines Tobias Pohlen Selected Topics in Human Language Technology and Pattern Recognition February 10, 2014 Human Language Technology and Pattern Recognition Lehrstuhl für Informatik 6
Support Vector Machines Tobias Pohlen Selected Topics in Human Language Technology and Pattern Recognition February 10, 2014 Human Language Technology and Pattern Recognition Lehrstuhl für Informatik 6
Pattern Recognition and Machine Learning. Perceptrons and Support Vector machines
 Pattern Recognition and Machine Learning James L. Crowley ENSIMAG 3 - MMIS Fall Semester 2016 Lessons 6 10 Jan 2017 Outline Perceptrons and Support Vector machines Notation... 2 Perceptrons... 3 History...3
Pattern Recognition and Machine Learning James L. Crowley ENSIMAG 3 - MMIS Fall Semester 2016 Lessons 6 10 Jan 2017 Outline Perceptrons and Support Vector machines Notation... 2 Perceptrons... 3 History...3
INFORMATION-THEORETIC BOUNDS OF EVOLUTIONARY PROCESSES MODELED AS A PROTEIN COMMUNICATION SYSTEM. Liuling Gong, Nidhal Bouaynaya and Dan Schonfeld
 INFORMATION-THEORETIC BOUNDS OF EVOLUTIONARY PROCESSES MODELED AS A PROTEIN COMMUNICATION SYSTEM Liuling Gong, Nidhal Bouaynaya and Dan Schonfeld University of Illinois at Chicago, Dept. of Electrical
INFORMATION-THEORETIC BOUNDS OF EVOLUTIONARY PROCESSES MODELED AS A PROTEIN COMMUNICATION SYSTEM Liuling Gong, Nidhal Bouaynaya and Dan Schonfeld University of Illinois at Chicago, Dept. of Electrical
Xia Ning,*, Huzefa Rangwala, and George Karypis
 J. Chem. Inf. Model. XXXX, xxx, 000 A Multi-Assay-Based Structure-Activity Relationship Models: Improving Structure-Activity Relationship Models by Incorporating Activity Information from Related Targets
J. Chem. Inf. Model. XXXX, xxx, 000 A Multi-Assay-Based Structure-Activity Relationship Models: Improving Structure-Activity Relationship Models by Incorporating Activity Information from Related Targets
Phylogenetic Networks, Trees, and Clusters
 Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Computational Genomics
 Computational Genomics http://www.cs.cmu.edu/~02710 Introduction to probability, statistics and algorithms (brief) intro to probability Basic notations Random variable - referring to an element / event
Computational Genomics http://www.cs.cmu.edu/~02710 Introduction to probability, statistics and algorithms (brief) intro to probability Basic notations Random variable - referring to an element / event
Outline. Basic concepts: SVM and kernels SVM primal/dual problems. Chih-Jen Lin (National Taiwan Univ.) 1 / 22
 Outline Basic concepts: SVM and kernels SVM primal/dual problems Chih-Jen Lin (National Taiwan Univ.) 1 / 22 Outline Basic concepts: SVM and kernels Basic concepts: SVM and kernels SVM primal/dual problems
Outline Basic concepts: SVM and kernels SVM primal/dual problems Chih-Jen Lin (National Taiwan Univ.) 1 / 22 Outline Basic concepts: SVM and kernels Basic concepts: SVM and kernels SVM primal/dual problems
Support Vector Machines and Kernel Algorithms
 Support Vector Machines and Kernel Algorithms Bernhard Schölkopf Max-Planck-Institut für biologische Kybernetik 72076 Tübingen, Germany Bernhard.Schoelkopf@tuebingen.mpg.de Alex Smola RSISE, Australian
Support Vector Machines and Kernel Algorithms Bernhard Schölkopf Max-Planck-Institut für biologische Kybernetik 72076 Tübingen, Germany Bernhard.Schoelkopf@tuebingen.mpg.de Alex Smola RSISE, Australian
Chapter 6: Classification
 Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
Bayesian Networks Inference with Probabilistic Graphical Models
 4190.408 2016-Spring Bayesian Networks Inference with Probabilistic Graphical Models Byoung-Tak Zhang intelligence Lab Seoul National University 4190.408 Artificial (2016-Spring) 1 Machine Learning? Learning
4190.408 2016-Spring Bayesian Networks Inference with Probabilistic Graphical Models Byoung-Tak Zhang intelligence Lab Seoul National University 4190.408 Artificial (2016-Spring) 1 Machine Learning? Learning
Introduction to Bioinformatics. Shifra Ben-Dor Irit Orr
 Introduction to Bioinformatics Shifra Ben-Dor Irit Orr Lecture Outline: Technical Course Items Introduction to Bioinformatics Introduction to Databases This week and next week What is bioinformatics? A
Introduction to Bioinformatics Shifra Ben-Dor Irit Orr Lecture Outline: Technical Course Items Introduction to Bioinformatics Introduction to Databases This week and next week What is bioinformatics? A
Mining Classification Knowledge
 Mining Classification Knowledge Remarks on NonSymbolic Methods JERZY STEFANOWSKI Institute of Computing Sciences, Poznań University of Technology COST Doctoral School, Troina 2008 Outline 1. Bayesian classification
Mining Classification Knowledge Remarks on NonSymbolic Methods JERZY STEFANOWSKI Institute of Computing Sciences, Poznań University of Technology COST Doctoral School, Troina 2008 Outline 1. Bayesian classification
Support Vector Machines for Classification: A Statistical Portrait
 Support Vector Machines for Classification: A Statistical Portrait Yoonkyung Lee Department of Statistics The Ohio State University May 27, 2011 The Spring Conference of Korean Statistical Society KAIST,
Support Vector Machines for Classification: A Statistical Portrait Yoonkyung Lee Department of Statistics The Ohio State University May 27, 2011 The Spring Conference of Korean Statistical Society KAIST,
#33 - Genomics 11/09/07
 BCB 444/544 Required Reading (before lecture) Lecture 33 Mon Nov 5 - Lecture 31 Phylogenetics Parsimony and ML Chp 11 - pp 142 169 Genomics Wed Nov 7 - Lecture 32 Machine Learning Fri Nov 9 - Lecture 33
BCB 444/544 Required Reading (before lecture) Lecture 33 Mon Nov 5 - Lecture 31 Phylogenetics Parsimony and ML Chp 11 - pp 142 169 Genomics Wed Nov 7 - Lecture 32 Machine Learning Fri Nov 9 - Lecture 33
The Decision List Machine
 The Decision List Machine Marina Sokolova SITE, University of Ottawa Ottawa, Ont. Canada,K1N-6N5 sokolova@site.uottawa.ca Nathalie Japkowicz SITE, University of Ottawa Ottawa, Ont. Canada,K1N-6N5 nat@site.uottawa.ca
The Decision List Machine Marina Sokolova SITE, University of Ottawa Ottawa, Ont. Canada,K1N-6N5 sokolova@site.uottawa.ca Nathalie Japkowicz SITE, University of Ottawa Ottawa, Ont. Canada,K1N-6N5 nat@site.uottawa.ca
6.047 / Computational Biology: Genomes, Networks, Evolution Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, etworks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, etworks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
BLAST: Target frequencies and information content Dannie Durand
 Computational Genomics and Molecular Biology, Fall 2016 1 BLAST: Target frequencies and information content Dannie Durand BLAST has two components: a fast heuristic for searching for similar sequences
Computational Genomics and Molecular Biology, Fall 2016 1 BLAST: Target frequencies and information content Dannie Durand BLAST has two components: a fast heuristic for searching for similar sequences
Support Vector Machine for Classification and Regression
 Support Vector Machine for Classification and Regression Ahlame Douzal AMA-LIG, Université Joseph Fourier Master 2R - MOSIG (2013) November 25, 2013 Loss function, Separating Hyperplanes, Canonical Hyperplan
Support Vector Machine for Classification and Regression Ahlame Douzal AMA-LIG, Université Joseph Fourier Master 2R - MOSIG (2013) November 25, 2013 Loss function, Separating Hyperplanes, Canonical Hyperplan
Polyhedral Computation. Linear Classifiers & the SVM
 Polyhedral Computation Linear Classifiers & the SVM mcuturi@i.kyoto-u.ac.jp Nov 26 2010 1 Statistical Inference Statistical: useful to study random systems... Mutations, environmental changes etc. life
Polyhedral Computation Linear Classifiers & the SVM mcuturi@i.kyoto-u.ac.jp Nov 26 2010 1 Statistical Inference Statistical: useful to study random systems... Mutations, environmental changes etc. life
GENE FUNCTION PREDICTION
 GENE FUNCTION PREDICTION Amal S. Perera Computer Science & Eng. University of Moratuwa. Moratuwa, Sri Lanka. shehan@uom.lk William Perrizo Computer Science North Dakota State University Fargo, ND 5815
GENE FUNCTION PREDICTION Amal S. Perera Computer Science & Eng. University of Moratuwa. Moratuwa, Sri Lanka. shehan@uom.lk William Perrizo Computer Science North Dakota State University Fargo, ND 5815
Algorithms in Bioinformatics FOUR Pairwise Sequence Alignment. Pairwise Sequence Alignment. Convention: DNA Sequences 5. Sequence Alignment
 Algorithms in Bioinformatics FOUR Sami Khuri Department of Computer Science San José State University Pairwise Sequence Alignment Homology Similarity Global string alignment Local string alignment Dot
Algorithms in Bioinformatics FOUR Sami Khuri Department of Computer Science San José State University Pairwise Sequence Alignment Homology Similarity Global string alignment Local string alignment Dot
Brief Introduction to Machine Learning
 Brief Introduction to Machine Learning Yuh-Jye Lee Lab of Data Science and Machine Intelligence Dept. of Applied Math. at NCTU August 29, 2016 1 / 49 1 Introduction 2 Binary Classification 3 Support Vector
Brief Introduction to Machine Learning Yuh-Jye Lee Lab of Data Science and Machine Intelligence Dept. of Applied Math. at NCTU August 29, 2016 1 / 49 1 Introduction 2 Binary Classification 3 Support Vector
Efficient Kernel Discriminant Analysis via QR Decomposition
 Efficient Kernel Discriminant Analysis via QR Decomposition Tao Xiong Department of ECE University of Minnesota txiong@ece.umn.edu Jieping Ye Department of CSE University of Minnesota jieping@cs.umn.edu
Efficient Kernel Discriminant Analysis via QR Decomposition Tao Xiong Department of ECE University of Minnesota txiong@ece.umn.edu Jieping Ye Department of CSE University of Minnesota jieping@cs.umn.edu
Single alignment: Substitution Matrix. 16 march 2017
 Single alignment: Substitution Matrix 16 march 2017 BLOSUM Matrix BLOSUM Matrix [2] (Blocks Amino Acid Substitution Matrices ) It is based on the amino acids substitutions observed in ~2000 conserved block
Single alignment: Substitution Matrix 16 march 2017 BLOSUM Matrix BLOSUM Matrix [2] (Blocks Amino Acid Substitution Matrices ) It is based on the amino acids substitutions observed in ~2000 conserved block
Linear Dependency Between and the Input Noise in -Support Vector Regression
 544 IEEE TRANSACTIONS ON NEURAL NETWORKS, VOL. 14, NO. 3, MAY 2003 Linear Dependency Between the Input Noise in -Support Vector Regression James T. Kwok Ivor W. Tsang Abstract In using the -support vector
544 IEEE TRANSACTIONS ON NEURAL NETWORKS, VOL. 14, NO. 3, MAY 2003 Linear Dependency Between the Input Noise in -Support Vector Regression James T. Kwok Ivor W. Tsang Abstract In using the -support vector
Statistical Learning and Kernel Methods in Bioinformatics
 Statistical Learning and Kernel Methods in Bioinformatics Bernhard Schölkopf, Isabelle Guyon, and Jason Weston Biowulf Technologies, New York Max-Planck-Institut für biologische Kybernetik, Tübingen, Biowulf
Statistical Learning and Kernel Methods in Bioinformatics Bernhard Schölkopf, Isabelle Guyon, and Jason Weston Biowulf Technologies, New York Max-Planck-Institut für biologische Kybernetik, Tübingen, Biowulf
Modern Information Retrieval
 Modern Information Retrieval Chapter 8 Text Classification Introduction A Characterization of Text Classification Unsupervised Algorithms Supervised Algorithms Feature Selection or Dimensionality Reduction
Modern Information Retrieval Chapter 8 Text Classification Introduction A Characterization of Text Classification Unsupervised Algorithms Supervised Algorithms Feature Selection or Dimensionality Reduction
Statistical learning theory, Support vector machines, and Bioinformatics
 1 Statistical learning theory, Support vector machines, and Bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group ENS Paris, november 25, 2003. 2 Overview 1.
1 Statistical learning theory, Support vector machines, and Bioinformatics Jean-Philippe.Vert@mines.org Ecole des Mines de Paris Computational Biology group ENS Paris, november 25, 2003. 2 Overview 1.
SVM Kernel Optimization: An Example in Yeast Protein Subcellular Localization Prediction
 SVM Kernel Optimization: An Example in Yeast Protein Subcellular Localization Prediction Ṭaráz E. Buck Computational Biology Program tebuck@andrew.cmu.edu Bin Zhang School of Public Policy and Management
SVM Kernel Optimization: An Example in Yeast Protein Subcellular Localization Prediction Ṭaráz E. Buck Computational Biology Program tebuck@andrew.cmu.edu Bin Zhang School of Public Policy and Management
O 3 O 4 O 5. q 3. q 4. Transition
 Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
Hidden Markov Models Hidden Markov models (HMM) were developed in the early part of the 1970 s and at that time mostly applied in the area of computerized speech recognition. They are first described in
Support Vector Machines for Protein Fold Class Prediction
 Florian Markowetz et al.: Support Vector Machines for Protein Fold Class Prediction Support Vector Machines for Protein Fold Class Prediction Florian Markowetz, Lutz Edler and Martin Vingron Department
Florian Markowetz et al.: Support Vector Machines for Protein Fold Class Prediction Support Vector Machines for Protein Fold Class Prediction Florian Markowetz, Lutz Edler and Martin Vingron Department
Final Exam, Machine Learning, Spring 2009
 Name: Andrew ID: Final Exam, 10701 Machine Learning, Spring 2009 - The exam is open-book, open-notes, no electronics other than calculators. - The maximum possible score on this exam is 100. You have 3
Name: Andrew ID: Final Exam, 10701 Machine Learning, Spring 2009 - The exam is open-book, open-notes, no electronics other than calculators. - The maximum possible score on this exam is 100. You have 3
