On stochastic models for the spread of infections. Jan Pieter Trapman
|
|
|
- Jade Shields
- 5 years ago
- Views:
Transcription
1 On stochastic models for the spread of infections Jan Pieter Trapman
2 c Pieter Trapman, De Bilt 2006 ISBN-10: ISBN-13: printed by PrintPartners Ipskamp, Enschede Engraving on cover: La peste di Firenze nel 1348
3 VRIJE UNIVERSITEIT On stochastic models for the spread of infections ACADEMISCH PROEFSCHRIFT ter verkrijging van de graad Doctor aan de Vrije Universiteit Amsterdam, op gezag van de rector magnificus, in het openbaar te verdedigen ten overstaan van de promotiecommissie van de faculteit der Exacte Wetenschappen op woensdag 13 september 2006 om uur in de aula van de universiteit, De Boelelaan 1105 door Jan Pieter Trapman geboren te Hardinxveld-Giessendam
4 promotoren: prof.dr. R.W.J. Meester prof.dr. J.A.P. Heesterbeek
5 I returned, and saw under the sun, that the race is not to the swift, nor the battle to the strong, neither yet bread to the wise, nor yet riches to men of understanding, nor yet favour to men of skill; but time and chance happeneth to them all. (Ecclesiastes 9:11, KJV)
6
7 Contents 1 General introduction Why mathematical epidemiology? Deterministic epidemic models Stochastic epidemic models Branching processes Percolation An overview of this thesis Possible future work in epidemiology List of publications A branching model for the spread of infectious animal diseases in varying environments Introduction Spread of the infection within a herd Spread of the infection between herds The model for varying environments Results Conclusion and final remarks Estimation in branching processes with restricted observations Introduction and motivation Formal set-up Results and application Estimating the offspring mean m Estimating a second function of the parameters Estimating a third function of parameters Conclusions Appendix
8 4 On pair approximation Introduction Set-up and notation The approximations Computation of basic quantities Remarks An example Random graphs as an approximation for networks on which infection spreads Introduction Notation and terminology Results from percolation theory Random networks Discussion Infection spread in a population of individuals with random infectivity and susceptibility Introduction Inhomogeneous random graphs: The model A comparison theorem for the random graphs with factorisable κ G (w, w) Discussion Bibliography 151 Samenvatting (in het Nederlands) 159
9 Chapter 1 General introduction 1.1 Why mathematical epidemiology? Recent outbreaks of animal diseases like classical swine fever (1997), foot and mouth disease (2001) and avian influenza (2003) in The Netherlands, the world-wide emerging of human diseases like SARS in 2002 and the constant threat of new pandemic infections, like avian influenza, make it needless to stress the importance of understanding and predicting the dynamics of the spread of an infection. Mathematical models and their analysis play a natural role in obtaining such understanding. Computer-intensive models can be used for predictions, but many of those models are mathematically intractable. Therefore, a challenge to epidemiologists, modellers and mathematicians is to find models that are both useful and mathematically tractable. The body of epidemiological literature is already immense. Although most of the models and methods cannot immediately be applied to epidemics in real-life, they do provide tools for the analysis of these real epidemics and give qualitative insight in the dynamics of the spread of the infection. In addition, real-life applications call for more advanced mathematical methods to be developed. Apart from the modelling of the spread of infections, results from epidemiology have been used in other sciences as well. One can think of models for rumour spread [26], the spread of information in economy [37], the spread of computer viruses [17, 31] and even an application of an epidemic model in a model on the spread of heresies in the Middle Ages [65]. 1
10 2 General introduction In this thesis I focus on the main purpose of mathematical epidemiology: modelling the spread of infections. Notably, I develop new methods to provide a better foundation for this modelling. The following sections of this general introduction are used for introducing some basic deterministic and stochastic epidemic theory and some theory of branching processes. Important epidemiological terms and quantities (like the basic reproduction ratio R 0 ) are introduced here. Readers familiar with the material can skip this part of the thesis without problems. After that a short introduction to the theory of percolation is given. This field in probability theory is mainly used in mathematical physics, but in this thesis an application of the theory in epidemiology is given. This chapter ends with an overview of this thesis and a section on possible directions for further development of (stochastic) mathematical epidemiology. 1.2 Deterministic epidemic models In 1927, 1932 and 1933 W.O. Kermack and A.G. McKendrick published a series of papers titled Contributions to the mathematical theory of epidemics. Those papers are often seen as the basis of further research in mathematical (especially deterministic) modelling of the spread of infectious diseases. The first three papers of Kermack and McKendrick are reprinted in [46]. In the book of Diekmann and Heesterbeek [27] some of the results of Kermack and McKendrick and other deterministic models are presented and explained. In many of the deterministic models randomly mixing populations are assumed, i.e. every individual contacts every other individual with the same rate, or in a multi-type process, contacts between individuals of certain types all happen at the same rate. First, I consider the basic single-type SIR (Susceptible Infectious Removed) epidemic model. Generalisation to multi-type models is often straightforward, but the notation becomes more cumbersome. In the model the population is closed and is of size N, i.e. births, deaths and migration are ignored. All pairs of individuals have contacts at the same rate N 1 β. If an infectious individual has a contact with a susceptible individual, the susceptible individual becomes infectious. Infectious individuals recover at rate α. Define S(t) as the number of individuals susceptible at time t and s(t) = N 1 S(t) as the fraction of the population that is susceptible at time t. I(t), i(t), R(t) and r(t) are defined similarly. Let i(0) = i 0 and s(0) = 1 i 0.
11 1.2 Deterministic epidemic models 3 This leads for large N to the following set of approximating differential equations: ds(t) dt di(t) dt dr(t) dt = βs(t)i(t), = βs(t)i(t) αi(t) = i(t)(βs(t) α), = αi(t). This is the place to introduce the concept of the basic reproduction ratio (or basic reproduction number) R 0. The basic reproduction ratio is defined as the number of secondary infections per initial infective individual, in a very large population where s(0) 1. In this model an initially infected individual will on average infect (N 1)β β N α α other individuals. So R 0 β/α. Some remarks have to be made: It can be seen from the differential equations that if a small number (relative to the total population size) of infective individuals is introduced in a completely susceptible population, the number of infectives will initially grow if β α > 0, i.e. if R 0 β α > 1. On the other hand, if R 0 1 the epidemic will go extinct very soon. This is the reason why R 0 is such an important quantity in epidemiology: It is a threshold parameter, like the offspring mean in branching processes (see Section 1.4). It is proven [27] that log(s( )) = R 0 (s( ) 1). This does not lead to a closed expression for s( ), but it can be computed numerically. Note that s( ) is the fraction of the total population that is never infected. Other deterministic models for infection spread usually lead to a comparable set of differential equations. In particular, it is easy to incorporate birth and death in the model. If birth and death are considered in the model we can also deduce conditions for an epidemic to become endemic, i.e. an epidemic for which i(t) 0 as t. A varying environment can be treated by making β and α time-dependent.
12 4 General introduction The duration of an epidemic cannot be studied in a realistic way with a deterministic model, because at the start and at the end of an epidemic stochastic fluctuations play a very important role. For some infections it is not enough to use three states of infection (susceptible, infective and removed), but we also need to take a latent state or more than one infectious states into account. One can deal with this by using a system of more differential equations than the three already given, but the main idea is still the same. The notion of the basic reproduction ratio R 0 is very important for controlling an epidemic. By vaccinating a fraction larger than 1 1 R 0 of a well mixed population at birth with a perfect vaccine, one can bring the reproduction ratio in the vaccinated population below 1 and thus prevent a major outbreak. An interesting problem is the modelling of spread of an infection among households. In this case two levels of infection are considered, the probability of infecting others within a household is higher than the probability of infecting individuals outside the own household. Note that for small households it is not very realistic to use a purely deterministic model. In this setting it is also very difficult to take a varying environment into account. The household model is also used in stochastic models [6], but in those models it is even more difficult to deal with varying environments (see Chapter 2). 1.3 Stochastic epidemic models Deterministic models have drawbacks and may cause some rather amusing mistakes: Murray et al. ([60], see also page 693 of [61]) proposed a deterministic model to predicted the dynamics of the prevalence of rabies among foxes in England. The dynamics predicted by the model seems to be rather spectacular. The number of infected foxes will rapidly increase until the number of available susceptible foxes is too low and then the disease seems to disappear, but after a period of about 2 years there is a sudden reappearing of rabies in foxes predicted. In 1991 Mollison [59] gave an explanation for this phenomenon, which he called the atto-fox phenomenon. Murray et al. have used a continuous approximation of the number of infected foxes (like is done in the differential equations in the deterministic SIR-model) and during the years
13 1.3 Stochastic epidemic models 5 that the infection seemed to have disappeared (in the predictions based on the model), there still was a minimum of around one infected atto-fox (10 18 of a fox) per square kilometre and this fox-part eventually caused the new wave of infection. This example shows the danger of continuous approximations of the number of individuals. Furthermore, as explained in the previous section for discrete models one has to assume that the number of infectious individuals is large and that the number of possible contacts with susceptible individuals is large as well. Therefore, deterministic models are not the most relevant for modelling the start and the end of an epidemic (where the number of infectious individuals is small) and for modelling the spread of infections on networks (where the number of available susceptible individuals that an infective individual can infect, is small). In this thesis we will mainly focus on these problematic cases i.e. we focus on stochastic and discrete models. Much of the work already done on stochastic epidemic models is described and explained in the lecture notes of Andersson and Britton [3]. In this section I give some of the results and the methods they describe The Reed-Frost model The most basic model is the Reed-Frost model. This model is generationbased. We assume that the population is closed, i.e. during the epidemic no new individuals are introduced in the population and there are no deaths. The probability of a susceptible to become infectious in generation i + 1 only depends on the number of infectives in generation i. We assume that the probability of escaping infection by a specific infective is q for each susceptible. Conditioned on the number of infectives in generation i the events that different susceptibles escape from the infection in generation i + 1 are independent. Note that in the last assumption, a fixed infective period and the same infectivity rate for all infectives is implicitly assumed. We get: P(I(j + 1) = n S(j) = k, I(j) = l) = (and S(j + 1) S(j) = I(j + 1)). ( ) k (1 q l ) n (q l ) k n n
14 6 General introduction We are interested in the final size (the number of initially susceptible individuals that are ultimately removed) of an epidemic. We can write this as: P(R( ) = k S(0) = n, I(0) = m) = l: l =k P(I(1) = l 1,..., I(j) = l j, I(j + 1) = 0 S(0) = n, I(0) = m), where l = j 1 l j. For practical purposes this formula is useless for large populations, because the needed number of computations grows very fast and worse: they are numerically unstable The standard stochastic SIR epidemic model For the stochastic SIR model we have the following assumptions: The population is closed, so no births, deaths and migration are considered in the model. Initially there are N m susceptibles and m infective individuals. The population is homogeneous and randomly mixing, i.e. the probability that two individuals contact each other does not depend on which two individuals we consider and all individuals have the same characteristics. At each contact between an infective and a susceptible the infection is transmitted. Each individual contacts a given other individual at the time points of a Poisson process with parameter β N. With some abuse of terminology β is called the infection rate or contact rate. An individual is in the R-class if its infectious period is terminated. Individuals have independent and identically distributed (i.i.d.) infectious periods, all distributed as the random variable I with mean ι and finite variance σ 2. The process described above is denoted by E N m,m (β, I). One remark on the terminology: from now on in this thesis the word rate in a stochastic setting will mean the density of a one dimensional Poisson process. The model proposed in this section slightly differs from models proposed in the literature (e.g. [3]). Andersson and Britton and many other authors
15 1.3 Stochastic epidemic models 7 working on stochastic epidemics assume that the contacts are made according β to a Poisson-process with parameter N m. In this model a process started with 2 infective and n susceptible individuals will behave in another way than a process initiated by 1 infective, and n + 1 susceptibles, given that one of the initially susceptible individuals is infected immediately. Note that replacing β by β(n m)/n in the model proposed in [3] brings us back at the standard stochastic SIR epidemic model described above. The basic reproduction number, R 0, is given by βι, i.e. the rate at which an infective individual contacts susceptible ones times the expected length of the infective period, or better: The expected number of secondary infected individuals directly infected by a single initially infected individual. We can look at the SIR model from a different perspective. Sellke gives such an alternative construction of the model [68]. In Sellke s construction each susceptible is associated with a threshold. As soon as the total infective pressure (i.e. β t N 0 I(u)du: the number of infective individuals times the infection rate integrated up to time t) is above the threshold the susceptible becomes infective and adds to the total infectivity pressure. More formally: The initial infectives are labelled: (m 1), (m 2),..., 0 and the initial susceptibles are labelled: 1, 2,..., n, where n = N m. We introduce random variables for the (possible) infectious periods, I j for (m 1) j n, all distributed as I. The initially susceptibles each get an exponential distributed random variable with mean one, Q j associated to it. The number of infectious individuals at time t is I(t). Let A(t) := β N t 0 I(u)du be the total infection pressure exerted on a given susceptible up to time t. Now the susceptible labeled j becomes infective as soon as A(t) is greater or equal than Q j. The j-th individual that becomes infected (not necessarily the susceptible labeled j) stays infective during a period I j, after this period it is removed. As soon as there are no more infectives the epidemic ceases. It is proved [3] that this model is equivalent to the SIR-model constructed earlier. Note that we can use this construction for the final size equation: { Z := R( ) m = min j Q (j+1) > β N j k= (m 1) I k }.
16 8 General introduction Here Q (j) denotes the j-th order statistic of the Q j s. pressure up to the end of the epidemic is now given by The total infection A := A( ) = β N Z k= (m 1) I k. (1.1) Andersson and Britton give the following results (first given by Ball in [4]): Lemma Consider the standard SIR epidemic E n,m (β, I) and let A be the total infection pressure of the epidemic. Then E[e θa /φ(βθ/n ) Z+m ] = 1, θ 0, where φ(θ) = E(e θi ) is the Laplace transform of I. This lemma, which is interesting on its own, is used to prove the following result: Theorem Consider the standard SIR epidemic E n,m (β, I). Denote by Pk n the probability that the final size of the epidemic is equal to k, 0 k n. Then l k=0 ( ) n k Pk n l k /[φ(β(n l)/n )]k+m = ( ) n, 0 l n. l With this theorem we can find the final size probabilities recursively. Note that we do not need to assume the absence of a latent period and we may replace β by β(τ), where τ is the time since infection of an individual. 1.4 Branching processes Two of the chapters (2 and 3) of this thesis have branching processes as their main subject, while in Chapter 5 basic theory from branching processes is heavily used to analyse infection spread on random networks. Indeed, there is a strong relation between epidemiology and the theory of branching processes, or to do more justice to the facts: mathematical epidemiology heavily relies on the theory of branching processes and draws inspiration from it. Branching processes were first developed to describe the extinction probabilities of family
17 1.4 Branching processes 9 names and can be seen as the mathematical study of family trees. (For an overview of the history of branching processes see the introduction of [39].) The basic (Galton-Watson) branching process model can roughly be seen as follows: There are some (male) ancestors that each have a random number of sons. The number of sons of the different ancestors are independently and identically distributed (i.i.d.). The sons of the ancestors have sons themselves, and the numbers of these sons are again i.i.d. and distributed as the offspring of an ancestor. In this way we can define a model for the whole family tree. All individuals have independently a number of sons, all according to the same distribution. The model can be enriched by considering the family tree in real time. In such an extended model not only the generation number of an individual is important, but also the times of birth and death are. Such a model is useful (and even needed) if, for example, one wants to make predictions about the number of people with surname Johnson in the year 2100 (Of course under the assumption that life expectancy, the distribution of the number of sons and the age at which people get their sons does not change between now and 2100). The relation between epidemics in large randomly mixing populations and family trees can heuristically be explained as follows. The individuals that introduce an infection into a population are seen as the ancestors. During their infectious period they contact a random number of individuals uniformly chosen from the population and infect those individuals if they are still susceptible. These newly infected individuals can be seen as generation 1 individuals. During their infectious period, these generation 1 individuals contact the other individuals according to the same law as the ancestors contacted other individuals. If the population is very large, the probability that an infective individual contacts an already infected individual during the first stage of the epidemic (which may last rather long) is very small, and therefore the progress of the epidemic can be described by a branching process. In the next subsection the so-called Galton-Watson processes are rigorously defined and some of the basic results on these processes are given. Furthermore, some extensions, like multi-type Galton-Watson processes, are defined and generalisations of the results on Galton-Watson processes are given. For proofs of the results and for further theory on branching processes one might
18 10 General introduction consult [39]. In subsection a theorem from [5] on the approximation of epidemics by branching processes is given. From now on we use the common convention of using the maternal terminology, so we speak of mothers that give birth to daughters Definition of the Galton-Watson process In this subsection we consider branching processes seen from a generation point of view, namely Galton-Watson processes. We assume that there is only one ancestor. This assumption is just for notational convenience and can easily be dropped. A Galton-Watson process is a process generating a family tree, where every individual has a finite (not necessarily bounded) number of daughters. The ancestor is labelled by a. The j n -th daughter of the j n 1 -th daughter... of the j 1 -th daughter of the ancestor is labelled by the vector (a, j 1,..., j n ). If an individual can give birth to more than one daughter at once, one can use a random ordering to label the daughters. The set of all possible labels J is given by J = {a {(a; x); x N n }}, n=1 where N is the set of positive integers (0 not included). This set of labels is countable. From now on individual x will mean the individual with label (a, x) J and individual a is the ancestor. Note that there are far too many labels, indeed if the ancestor has only 1 daughter, there are no individuals with the labels of the form (a, 2, j 2, j n ; n N, j i N). Let r x be 1 if the individual x is realised, i.e. r x = 1 if the label (a, x) is assigned to an individual. Otherwise r x = 0. Let {ξ x, x J} be a set of random variables that are i.i.d. and distributed as a random variable ξ. If r x = 1, then ξ x denotes the total number of daughters of individual x. Furthermore, let p k := P(ξ = k).
19 1.4 Branching processes 11 Now define the random variables X n for n N, X n := x N n r x = x N n 1 r x ξ x, as the total number of individuals in generation n. Let X 0 be the number of ancestors, which we have assumed to be 1. {X n ; n 0 N} form a homogeneous Markov chain The generating function and the extinction probability The (probability) generating function of a discrete random variable R [32, 39] is defined as f R (s) := E(s R ) = P(R = i)s i for all s R 0 for which this sum converges. If s 1 the sum converges. The generating function has useful properties like i=0 f R(1) = E(R), f R(1) = E(R(R 1)), f (k) R (0) = k!p(r = k), where f (k) R (s) is the k-th derivative of f R(s) and f R (1) = lim s 1 f R (s) if the generating function is not defined for s > 1. The last property shows that f R (s) determines the distribution of R. For the analysis of Galton-Watson processes we consider the following generating functions for k N. f(s) := f ξ (s) = E(s ξ ), f k (s) := f Xk (s) = E(s X k )
20 12 General introduction Note that f k+1 (s) = E(s X k+1 ) = E(E(s X k+1 X k )) = E([E(s ξ )] X k ) = E([f(s)] X k ) = f k (f(s)) and by induction we obtain f k+1 (s) = f(f k (s)). The generating function is extremely important in the theory of Galton- Watson processes, because it is the key to finding the extinction probability of the process, and it is the quest for extinction probabilities that led to the development of the theory of branching processes. Some simple arguments can explain the relation between the generating function and the extinction probability. Let q be the probability that the progeny of a single ancestor goes extinct. If the progeny of the ancestor goes extinct this means that she has no daughters at all or that the progeny of all of her daughters goes extinct. However, the number of daughters of different individuals are i.i.d. This brings us to the equation q = p i q i = E(q ξ ) = f(q). (1.2) i=0 This argument has been used in Chapter 2 to obtain equations to compute the probability of a major outbreak of an epidemic. If the offspring mean m := E(ξ) is larger than 1, then the equation above has two roots in [0, 1]. In that case, it is well known [39] that the extinction probability is the smallest of the two. The other root is always 1. If m 1 and p 1 1, then 1 is the only root in [0, 1]. Galton-Watson processes with m = 1 are called critical processes, and if m is greater or less than 1 the processes are respectively supercritical and subcritical. An important property of Galton-Watson processes is that with probability 1 they either go extinct or explode, i.e. with probability 1, lim k X k is either 0 or (Theorem of [39]). In this thesis I mainly consider supercritical branching processes. In Chapter 2 the extinction probability of a special branching process is computed. In
21 1.4 Branching processes 13 Chapter 3 moments of the offspring distribution are estimated. The estimators only converge on the set where the process does not go extinct. In Chapter 5 we use branching processes to determine features like the reproduction ratio R (a quantity similar to R 0, which in turn can be interpreted as the offspring mean m) and the extinction probability for the spread of an infection on random networks Martingale convergence Martingales play an important role in the theory of branching processes, especially for proving convergence results one can hardly do without. This becomes apparent in Chapter 3 where we make frequent use of martingales in order to prove convergence of estimators for the moments of the offspring distribution. The theorems used is Chapter 3 indirectly use the following basic theorem from Branching processes (compare this with Theorems and of [39]), Theorem If E(ξ log(ξ)) < the martingale W n := m n X n converges almost surely and in L 1 norm to a non-degenerate random variable W, which is 0 if and only if X n Multi-type Galton-Watson processes In this subsection some results from the theory of multi-type Galton-Watson branching processes are given (See Chapter 4 of [39]). We consider processes with only one ancestor. Results can easily be generalised to processes with more ancestors. There exist results similar to those of single-type branching processes for r-type Galton-Watson processes. Let X n (i) be the vector with the number of individuals of the different types in generation n, given that the ancestor is of type i. For 1 i, j r, let m ij be the mean number of j-daughters of one i-individual (i.e. m ij = E[(X 1 (i)) j ]). This defines an r r matrix m. We assume that the matrix m is positively regular, i.e. there exists an n > 0 such that all entries in m n are strictly positive. Let ρ be the positive eigenvalue of m which is greater than or equal to all other eigenvalues in absolute value (this eigenvalue exists by the Perron-Frobenius theorem for irreducible matrices). Furthermore we use the following vector notation.
22 14 General introduction s = [s 1,..., s r ] [0, 1] r, k = [k 1,... k r ] Z r +, 0 = [0,..., 0] Z r +, f (i) (s) = k P((X 1 (i)) j = k j ; 1 j r)s k skr r for 1 i r, f(s) = [f (1) (s),..., f (r) (s)]. Let q i be the probability of extinction of the branching process given that it started with one i individual. From Theorem of [39] we know that the [q 1,..., q r ] is the solution of f(s) = s with the smallest Euclidean distance to the origin. If ρ > 1, then q i < 1 for all i and if ρ 1, then q i = 1 for all i. Furthermore, Theorem of [39] states that if ρ > 1 then ρ n X n (k) W k v, where v is the left eigenvector of m and W k is a random variable with the property that {W k > 0} differs from {lim n X n (k) 0} only on a null-set General branching processes Galton-Watson processes are used to describe a family tree from the perspective of generations. However in epidemiology one may be more interested in the real time development of the number of infective individuals. Usually it is not possible to observe the size of an infection generation, but it may be possible to observe the number of infective individuals at a certain time. The theory of branching processes goes further than only Galton-Watson processes, and it is possible to define a branching process in real time. However, results are much harder to obtain. The process is defined as follows. Assign i.i.d. a pair (λ x, η x ) to every label x J. If x is realised in the branching process, the non negative random variable λ x is the life length of individual x, while η x is a point process on the non-negative real line denoting the reproduction of individual x. Let η x ([t 1, t 2 ]) denote the number of points of η x in the interval [t 1, t 2 ] and use η x (t) as a shorthand for η x ([0, t]). The total number of daughters of x is given by η x (λ x ) = η x ( ). From now on, we drop the subscript x, if it does not lead to confusion. We define µ([t 1, t 2 ]) := E(η([t 1, t 2 ])) and µ(t) := E(η(t)) < 1. For technical reasons we assume that the process is non-lattice i.e. there is no d > 0 such that k=0 µ({kd}) = µ( ). Furthermore, we assume that µ( ) < and µ(0) < 1. It is proven [39] that this
23 1.4 Branching processes 15 last assumption is enough to guarantee that at any given time the number of living individuals is finite. We are interested in the number of living individuals younger than a at time t, X a t. The following theorem from [39] can be used: Theorem For a general, non-lattice branching process with µ( ) > 1 and V ar[η( )] <, let α be the Malthusian parameter, i.e. the root of It holds that lim t e αt Xt a written as lim t e αt Xt a = Z a 0 0 e αt µ(dt) = 1. exists a.s. and in mean square. The limit can be e αt P[λ > t]dt/β = Z{1 E[e α(a t) ]}/(αβ), where β = 0 te αt µ({dt}) is the mothers average age of child bearing and Z is a random variable such that E[Z] = 1 and P[Z = 0, X t 0] = Approximating epidemics by branching processes The start of an SIR epidemic in a large population can be approximated by the start of a continuous time branching process. Ball and Donnelly give some useful results in [5]. I give one of those results here: Theorem There is a probability space (Ω, F, P) on which are defined a sequence of epidemic models indexed by n (the initial number of susceptibles) and the approximating branching process, with the following properties: Let I n (t) be the number of infectives at time t in a population of size n. And X(t) the number of individuals in the branching process at time t. Denote by A the set on which the branching process X( ) does not go extinct: A = { } ω Ω : lim X(t, ω) 0. t We use A c to denote the complement of A. Then, as n, for P almost all ω A c. sup I n (t) X(t) 0 0 t
24 16 General introduction Further for any c 1 < (2α) 1 and c 2 > (2α) 1, sup I n (t) X(t) 0 0 t c 1 log n and sup I n (t) X(t) 0 t c 2 log n as n, for P almost all ω A. Here α is the Malthusian parameter and µ(t) is the expected number of children of an ancestor of the branching process up to time t as before. This theorem implies that if the initial number of susceptible individuals goes to infinity, then the probability of a small outbreak in the population converge to the probability of extinction in the corresponding branching process. Furthermore, if there is a small outbreak, then the distribution of the final size of the epidemic is in the limit equal to the distribution of the final size of the corresponding branching process on the extinction set. Finally it is shown that epidemic processes are well described by branching processes up to a time of order log n. 1.5 Percolation Consider a very large orchard with trees planted at regular distances in such a way that the positions of the trees can be seen as the vertices of the square lattice. Now assume that one of the trees somewhere near the centre of the orchard is infected by a disease. The infection has the following characteristics. Exactly one time unit after being infected a tree will die. As long as a tree is infectious (i.e. until its death) it will spread infectious material to its nearest neighbours, which thereby become infected if they were not already so. The probability that a given nearest neighbour receives infectious material from the infectious tree is p and infections of nearest neighbours happen independently of each other. No infectious material is spread farther than the nearest neighbours. Note that the infection under consideration has SIR dynamics, where the R state stands for death. To model the spread of such an infection on a square lattice we use percolation theory. (For a rigorous and extensive treatment of the subject see [33]). In the percolation model the dynamic character of the
25 1.5 Percolation 17 epidemic spread has been dropped and only the static clusters of the spread are considered The model The network that is mainly studied in percolation theory is the graph where the vertices (sites, nodes) are the points of the regular lattice Z d and edges (bonds, connections) are drawn between any two vertices at Euclidean distance 1. However, other networks can be studied as well. Vertices connected by an edge are neighbours. Edges are open with probability p independently of each other. If an edge is not open then it is closed. Examples of question to be answered are: Is there a positive probability that one can reach infinitely many vertices from the origin by crossing only open edges? (i.e. is the origin contained in an infinite open cluster?) How does this probability depend on p? Does there exists a value p c, strictly between 0 and 1, with the property that the probability the origin is part of an infinite open cluster is 0 for p < p c and strictly larger than 0 if p > p c? The model described above is called bond percolation. We can also consider site percolation, where vertices are open with probability p independently of each other. The open cluster of the origin here consists of the vertices that can be reached by paths consisting of edges with two open end-vertices. In more mathematical notation we use the measure P p for the measure associated to the bond percolation model described above. We drop the subscript if we are not immediately interested in how the probability of an event dependent on p. The probability that the origin is part of an infinite cluster is denoted by θ(p) := P p (0 ) and p c := inf{p : θ(p) > 0}. Kesten [45] proved that for bond percolation on Z 2 the critical probability is given by p c = 1/2. For site percolation p c on the square lattice is unknown. In fact we should have mentioned the underlying graph in the above notation as well, but in general it is clear which graph we consider. Note that we are using product measure, so generalisations to other graphs are straightforward. For epidemiological purposes it is more natural to consider a directed underlying graph. In such a graph edges are replaced by directed edges. The presence of an edge from vertex v 1 to vertex v 2 does not necessarily imply that there is an edge from v 2 to v 1, although in some models (like the one presented in Chapter 5) only graphs where this implication holds, are considered. It is
26 18 General introduction still the product measure that is considered, so whether or not the edge from v 1 to v 2 is open is independent of the state of the other edges, in particular it is independent of the state of the edge from v 2 to v 1. The cluster of the origin consists of those vertices that can be reached by an open path from the origin A relation between percolation and SIR epidemics In this subsection we consider undirected graphs. One can explore the static (bond) percolation cluster of the origin in the following dynamic way. The construction of a percolation cluster on directed graphs is similar to the construction below. Say that the origin is the generation 0 vertex. Now explore the states (open or closed) of all the edges with the origin as one of the endpoints. The other endpoints of the explored open edges are the generation 1 vertices. The following step is to explore all the edges with a generation 1 vertex as endpoint, that were not explored before. The endpoints of the newly explored open edges are the generation 2 vertices, if there was no other generation number assigned to it yet. If a vertex has a generation number assigned to it, it keeps that number for ever. The n+1-th step is to explore the states of all the edges with a generation n vertex as end vertex, that were not explored before, and say that all the end vertices of the open edges that had not assigned a generation number to it yet, are generation n+1 vertices. This procedure will stop in a finite number of steps if and only if the cluster of the origin is finite. Now return to the example of the orchard above and explore whether or not infectious material is transmitted between a pair of trees. First explore the trees next to the initial infective tree. If infectious material is transmitted to such a tree from the initial infective tree, it is said to be a generation 1 tree. Now explore whether or not infectious material is transmitted from the generation 1 trees to its neighbours for the pairs that have not been explored before. In such a way the whole cluster of infected trees can be explored, and in fact this exploration procedure is exactly the same as the exploration procedure of the percolation cluster. So the probability of an infinite outbreak of the tree-infection in an infinite orchard is θ(p). In particular, if the probability that a tree transmits infectious material to a given neighbouring tree is larger than 0.5, there is a positive probability of a large outbreak of the infection.
27 1.5 Percolation 19 Here we use that it does not matter for the size of the cluster whether or not the edges are directed. Because the second endpoint of an edge of which the corresponding edge in opposite direction has already been explored is not susceptible anymore. The exploration procedure can easily be generalised to more than one (but finitely many) initial infectives on more general graphs that are locally finite (i.e. graphs where each vertex has a finite number of neighbours) Locally dependent percolation The main extension of percolation theory that is used in this thesis, is locally dependent percolation by which locally dependent random graphs are constructed (see Chapter 5 and [25, 51]). We consider percolation on a directed graph, where each vertex has a finite number of edges starting at it. We introduce randomness at two levels. First we assign infectious periods I(v) independently and identically distributed to each vertex. The second step is declaring the edges in the graph open or closed again using product measure, but in this model the edge from v 1 to v 2 is open with probability p(i(v 1 )) = 1 exp[ τi(v 1 )], for some infection rate τ. By using the terms infectious period and infection rate we already gave away the relation with epidemic spread. We need this extension because in real-life response to an infection may differ between individuals. One of the things that may be different is the period that an individual stays infectious. In the epidemic model corresponding with locally dependent percolation, these infectious periods are assumed to be i.i.d. Now define p := E(1 e τi ) as the marginal probability that a given bond is open. Kuulasmaa [51] proved that if we compare locally dependent percolation with the same marginal probabilities that edges are open, a fixed infectious period gives a worst case scenario, in the sense that the probability of the origin being in an infinite cluster is maximal. This model corresponds with bond-percolation. In [25] it is shown that the process where all edges going out of a vertex are open with probability p and all of them are closed with probability 1 p gives a best-case scenario, in the sense that the probability of the origin being in an infinite cluster is minimal.
28 20 General introduction 1.6 An overview of this thesis The goal of the research presented in this thesis is the development of new stochastic epidemic models and methods, to extend the existing methods and to apply models and methods from other areas of probability theory or more general mathematics to epidemiology. When I started the project The Netherlands had just been hit by several outbreaks of infectious diseases of animals, that have had great impact on animal health and welfare and the economy of the country. During the outbreaks the government and the farmers took measures to stop the epidemic and to save animals. One of the effects of the measures is that the dynamics of the spread changes during the outbreak itself. Of course this is exactly what the measures are for: reducing the probability that the infection is transmitted from an infected farm to a susceptible one. However, this change in the dynamics is hard to incorporate in the existing models in epidemiology. In Chapter 2 a model is developed that can be used to study the real-time dynamics of the spread of an infection in varying environments. This model has been used to analyse the effects of various control measures. The model proposed in Chapter 2 is mainly on branching processes in varying environments. In that branching model properties of the infection and the parameters that describe the spread of the infection in a non-varying environment are used and it is assumed that these parameters are known. In Chapter 3 the leading question is whether we can obtain the needed parameters from a branching process if we only have restricted observations? It is proven in [34] that if we only observe the generation sizes, then we can only estimate two parameters of the branching process consistently. This means that even if we observe the process for an infinite period of time, we can only be sure about two of the parameters. In our case we do not observe the generation sizes, but only the individuals that stop being in the process (the farms at which an infection is detected). Surprisingly, with these observations it is possible to estimate three parameters consistently. One drawback of the approach is that two of the three estimators converge very slowly and for practical purposes much too slow. One of the conclusions drawn in this chapter is that more information is needed than only the detection times of the farm in order to estimate the parameters in the branching model of infection spread.
29 1.6 An overview of this thesis 21 In Chapters 2 and 3 real-time processes are studied in large randomly mixing populations. However, for some purposes the assumption of randomly mixing populations is strong and seems to be unrealistic. In Chapters 4, 5 and 6 methods are provided to study the spread of infections on networks. However, the cost of this extra structure is that in the Chapters 5 and 6 we loose the real-time perspective on the epidemic. In the three chapters on infection spread on networks some of the properties of the (social) network on which the infection spreads are captured in the models. In Chapter 4 the method of pair-approximations is discussed. This method is used in deterministic models and it can roughly be seen as a model of randomly mixing pairs of individuals in stead of randomly mixing single individuals. Although the intuition behind this model seems to be clear, it is hard to make all the assumptions and approximations, usually used in pair approximation models, explicit. In this thesis I look at the approximation from the viewpoint of a probabilist. The model assumptions are made explicit and the approximations used in the existing models are analysed. Furthermore, the dynamics of the expectation of the number of infected individuals are described instead of assuming that this expectation is the same as the actual number of individuals (which is the assumption in most of the deterministic models in epidemiology). Furthermore a new proposal for a useful reproduction number is made. In Chapter 4 the time dynamics of the epidemic are studied. However keeping track of this time dynamics makes it necessary to make some extra assumptions. In Chapter 5 the real-time perspective is replaced by a generation perspective, where we do not consider the times at which infections took place, but instead of that we use the infection tree, where connections are the actual infections that took place. Note that for quantities like the probability of a large outbreak and the expected number of individuals that will ultimately be infected, the perspective does not matter. Still, mathematical analysis of the spread of an infection on a general network is far too difficult, therefore we replace the general network by a random network which has some features in common with the original network. We determine a reproduction number and the probability of a major outbreak on the random network. Replacing networks by random networks for modelling purposes is not new. However, in the existing models on epidemics on networks, the random network that is used has no or only few triangles in it. In real-life triangles
30 22 General introduction naturally arise in social networks (the friends of my friends are often also my friends). In Chapter 5 random networks are constructed that do have an a priori given expected number of triangles. Finally, in Chapter 6 the relation between a certain type of random networks (the generalised random graphs) and epidemic spread is established and results known from percolation theory (especially the results of Kuulasmaa [51], see Section 1.5) are generalised to these random graphs in order to give worst case scenarios for epidemics in heterogeneous populations. The generalised random graphs are well fit to model the spread of an infection in a heterogeneous population, where individuals have random infectivity and susceptibility (the weights of the individuals). This chapter can be seen as an ansatz for further research. 1.7 Possible future work in epidemiology As mentioned above, Chapter 6 gives a foundation for further work on modelling the spread of infections in heterogeneous populations. Especially the important question on the population effect of vaccines which do not give total protection and which cause a change in infectivity, can be tackled. Other future work can be expected from the field of percolation theory Long-range percolation The model described in Section 1.5 is elegant, but it does not give a very realistic description of the spread of an infection. A possible extension of the model is long-range percolation. This model has not directly been used in this thesis, but it is useful to compare this section with the generalised random graphs of Chapter 6. While edges in generalised random graphs are open based on the weights of the vertices, in long-range percolation they are open based on the distance between two vertices. We change the terminology a bit by replacing open by present and closed by absent. Using this terminology makes it clearer that we are actually constructing a random graph. The construction is done as follows: We start with a vertex set V in some metric space. For simplicity we assume V = Z 2 or more general V = Z d. For each pair of vertices (v, w) there is an edge joining them with probability p( v w ), where v w denotes the Euclidian distance (any other metric will
31 1.7 Possible future work in epidemiology 23 do as well). The presence or absence of an edge is independent of the presence or absence of other edges, so we still use product measure. It is natural to assume that p(x) is a decreasing function. Long-range percolation provides a much richer way of modelling the spread of infections than ordinary percolation. Of course analysis becomes much harder as well. In general it is very hard to answer questions on extinction probabilities. However, questions that are little studied in ordinary percolation arise in long-range percolation. One of those questions is concerned the diameter of large subgraphs of Z d. Consider the block B N := [0, N] d Z d. Edges are added according to the rules of long-range percolation. The random graph obtained in this way is called G. Define for v, w B N, D(v, w) as the graph distance or chemical distance between v and w, i.e. the minimal number of edges in G that has to be crossed to go from v to w. If there is no path from v to w the chemical distance is said to be infinite. The diameter D N is defined as D N := max D(v, w). (1.3) v,w B N In general one restricts attention to a connected component of the graph, because D N = is not informative. Results on D N and D(v, w) for different regimes of p(x) are given in [15, 16, 18, 24]. Marek Biskup proved the following theorem: Theorem Consider long-range percolation on Z d. Let s (d, 2d). Let p(x) = 1 exp[ q(x)], with log q(x) lim = s (1.4) x log x and assume that, the random graph G a.s. contains a unique infinite component C. Then for all ɛ > 0, ( lim P log D(0, x) ) ɛ + ɛ 0, x C = 1, (1.5) x log log x where = (s, d) = log 2 log(2d/s). (1.6)
Reproduction numbers for epidemic models with households and other social structures I: Definition and calculation of R 0
 Mathematical Statistics Stockholm University Reproduction numbers for epidemic models with households and other social structures I: Definition and calculation of R 0 Lorenzo Pellis Frank Ball Pieter Trapman
Mathematical Statistics Stockholm University Reproduction numbers for epidemic models with households and other social structures I: Definition and calculation of R 0 Lorenzo Pellis Frank Ball Pieter Trapman
Threshold Parameter for a Random Graph Epidemic Model
 Advances in Applied Mathematical Biosciences. ISSN 2248-9983 Volume 5, Number 1 (2014), pp. 35-41 International Research Publication House http://www.irphouse.com Threshold Parameter for a Random Graph
Advances in Applied Mathematical Biosciences. ISSN 2248-9983 Volume 5, Number 1 (2014), pp. 35-41 International Research Publication House http://www.irphouse.com Threshold Parameter for a Random Graph
Three Disguises of 1 x = e λx
 Three Disguises of 1 x = e λx Chathuri Karunarathna Mudiyanselage Rabi K.C. Winfried Just Department of Mathematics, Ohio University Mathematical Biology and Dynamical Systems Seminar Ohio University November
Three Disguises of 1 x = e λx Chathuri Karunarathna Mudiyanselage Rabi K.C. Winfried Just Department of Mathematics, Ohio University Mathematical Biology and Dynamical Systems Seminar Ohio University November
Modern Discrete Probability Branching processes
 Modern Discrete Probability IV - Branching processes Review Sébastien Roch UW Madison Mathematics November 15, 2014 1 Basic definitions 2 3 4 Galton-Watson branching processes I Definition A Galton-Watson
Modern Discrete Probability IV - Branching processes Review Sébastien Roch UW Madison Mathematics November 15, 2014 1 Basic definitions 2 3 4 Galton-Watson branching processes I Definition A Galton-Watson
LAW OF LARGE NUMBERS FOR THE SIRS EPIDEMIC
 LAW OF LARGE NUMBERS FOR THE SIRS EPIDEMIC R. G. DOLGOARSHINNYKH Abstract. We establish law of large numbers for SIRS stochastic epidemic processes: as the population size increases the paths of SIRS epidemic
LAW OF LARGE NUMBERS FOR THE SIRS EPIDEMIC R. G. DOLGOARSHINNYKH Abstract. We establish law of large numbers for SIRS stochastic epidemic processes: as the population size increases the paths of SIRS epidemic
Reproduction numbers for epidemic models with households and other social structures II: comparisons and implications for vaccination
 Reproduction numbers for epidemic models with households and other social structures II: comparisons and implications for vaccination arxiv:1410.4469v [q-bio.pe] 10 Dec 015 Frank Ball 1, Lorenzo Pellis
Reproduction numbers for epidemic models with households and other social structures II: comparisons and implications for vaccination arxiv:1410.4469v [q-bio.pe] 10 Dec 015 Frank Ball 1, Lorenzo Pellis
SIR epidemics on random graphs with clustering
 SIR epidemics on random graphs with clustering Carolina Fransson Masteruppsats i matematisk statistik Master Thesis in Mathematical Statistics Masteruppsats 2017:3 Matematisk statistik Maj 2017 www.math.su.se
SIR epidemics on random graphs with clustering Carolina Fransson Masteruppsats i matematisk statistik Master Thesis in Mathematical Statistics Masteruppsats 2017:3 Matematisk statistik Maj 2017 www.math.su.se
Stochastic modelling of epidemic spread
 Stochastic modelling of epidemic spread Julien Arino Centre for Research on Inner City Health St Michael s Hospital Toronto On leave from Department of Mathematics University of Manitoba Julien Arino@umanitoba.ca
Stochastic modelling of epidemic spread Julien Arino Centre for Research on Inner City Health St Michael s Hospital Toronto On leave from Department of Mathematics University of Manitoba Julien Arino@umanitoba.ca
Partially Observed Stochastic Epidemics
 Institut de Mathématiques Ecole Polytechnique Fédérale de Lausanne victor.panaretos@epfl.ch http://smat.epfl.ch Stochastic Epidemics Stochastic Epidemic = stochastic model for evolution of epidemic { stochastic
Institut de Mathématiques Ecole Polytechnique Fédérale de Lausanne victor.panaretos@epfl.ch http://smat.epfl.ch Stochastic Epidemics Stochastic Epidemic = stochastic model for evolution of epidemic { stochastic
4: The Pandemic process
 4: The Pandemic process David Aldous July 12, 2012 (repeat of previous slide) Background meeting model with rates ν. Model: Pandemic Initially one agent is infected. Whenever an infected agent meets another
4: The Pandemic process David Aldous July 12, 2012 (repeat of previous slide) Background meeting model with rates ν. Model: Pandemic Initially one agent is infected. Whenever an infected agent meets another
Chapter 6: Branching Processes: The Theory of Reproduction
 116 Chapter 6: Branching Processes: The Theory of Reproduction Viruses Aphids Royalty DNA Although the early development of Probability Theory was motivated by problems in gambling, probabilists soon realised
116 Chapter 6: Branching Processes: The Theory of Reproduction Viruses Aphids Royalty DNA Although the early development of Probability Theory was motivated by problems in gambling, probabilists soon realised
Mathematical Epidemiology Lecture 1. Matylda Jabłońska-Sabuka
 Lecture 1 Lappeenranta University of Technology Wrocław, Fall 2013 What is? Basic terminology Epidemiology is the subject that studies the spread of diseases in populations, and primarily the human populations.
Lecture 1 Lappeenranta University of Technology Wrocław, Fall 2013 What is? Basic terminology Epidemiology is the subject that studies the spread of diseases in populations, and primarily the human populations.
Thursday. Threshold and Sensitivity Analysis
 Thursday Threshold and Sensitivity Analysis SIR Model without Demography ds dt di dt dr dt = βsi (2.1) = βsi γi (2.2) = γi (2.3) With initial conditions S(0) > 0, I(0) > 0, and R(0) = 0. This model can
Thursday Threshold and Sensitivity Analysis SIR Model without Demography ds dt di dt dr dt = βsi (2.1) = βsi γi (2.2) = γi (2.3) With initial conditions S(0) > 0, I(0) > 0, and R(0) = 0. This model can
Invariant measures and limiting shapes in sandpile models. Haiyan Liu
 Invariant measures and limiting shapes in sandpile models Haiyan Liu c H. Liu, Amsterdam 2011 ISBN: 9789086595259 Printed by Ipskamp Drukkers, Enschede, The Netherlands VRIJE UNIVERSITEIT Invariant measures
Invariant measures and limiting shapes in sandpile models Haiyan Liu c H. Liu, Amsterdam 2011 ISBN: 9789086595259 Printed by Ipskamp Drukkers, Enschede, The Netherlands VRIJE UNIVERSITEIT Invariant measures
Course Notes. Part IV. Probabilistic Combinatorics. Algorithms
 Course Notes Part IV Probabilistic Combinatorics and Algorithms J. A. Verstraete Department of Mathematics University of California San Diego 9500 Gilman Drive La Jolla California 92037-0112 jacques@ucsd.edu
Course Notes Part IV Probabilistic Combinatorics and Algorithms J. A. Verstraete Department of Mathematics University of California San Diego 9500 Gilman Drive La Jolla California 92037-0112 jacques@ucsd.edu
Copyright is owned by the Author of the thesis. Permission is given for a copy to be downloaded by an individual for the purpose of research and
 Copyright is owned by the Author of the thesis. Permission is given for a copy to be downloaded by an individual for the purpose of research and private study only. The thesis may not be reproduced elsewhere
Copyright is owned by the Author of the thesis. Permission is given for a copy to be downloaded by an individual for the purpose of research and private study only. The thesis may not be reproduced elsewhere
6.207/14.15: Networks Lecture 12: Generalized Random Graphs
 6.207/14.15: Networks Lecture 12: Generalized Random Graphs 1 Outline Small-world model Growing random networks Power-law degree distributions: Rich-Get-Richer effects Models: Uniform attachment model
6.207/14.15: Networks Lecture 12: Generalized Random Graphs 1 Outline Small-world model Growing random networks Power-law degree distributions: Rich-Get-Richer effects Models: Uniform attachment model
Chapter 11. Min Cut Min Cut Problem Definition Some Definitions. By Sariel Har-Peled, December 10, Version: 1.
 Chapter 11 Min Cut By Sariel Har-Peled, December 10, 013 1 Version: 1.0 I built on the sand And it tumbled down, I built on a rock And it tumbled down. Now when I build, I shall begin With the smoke from
Chapter 11 Min Cut By Sariel Har-Peled, December 10, 013 1 Version: 1.0 I built on the sand And it tumbled down, I built on a rock And it tumbled down. Now when I build, I shall begin With the smoke from
A branching model for the spread of infectious animal diseases in varying environments
 J. Math. Biol. 49: 553 576 (2004) Mathematical Biology Digital Object Identifier (DOI): 10.1007/s00285-004-0267-5 Pieter Trapman Ronald Meester Hans Heesterbeek A branching model for the spread of infectious
J. Math. Biol. 49: 553 576 (2004) Mathematical Biology Digital Object Identifier (DOI): 10.1007/s00285-004-0267-5 Pieter Trapman Ronald Meester Hans Heesterbeek A branching model for the spread of infectious
1. Let X and Y be independent exponential random variables with rate α. Find the densities of the random variables X 3, X Y, min(x, Y 3 )
 1 Introduction These problems are meant to be practice problems for you to see if you have understood the material reasonably well. They are neither exhaustive (e.g. Diffusions, continuous time branching
1 Introduction These problems are meant to be practice problems for you to see if you have understood the material reasonably well. They are neither exhaustive (e.g. Diffusions, continuous time branching
EPIDEMICS WITH TWO LEVELS OF MIXING
 The Annals of Applied Probability 1997, Vol. 7, No. 1, 46 89 EPIDEMICS WITH TWO LEVELS OF MIXING By Frank Ball, Denis Mollison and Gianpaolo Scalia-Tomba University of Nottingham, Heriot-Watt University
The Annals of Applied Probability 1997, Vol. 7, No. 1, 46 89 EPIDEMICS WITH TWO LEVELS OF MIXING By Frank Ball, Denis Mollison and Gianpaolo Scalia-Tomba University of Nottingham, Heriot-Watt University
Introduction to SEIR Models
 Department of Epidemiology and Public Health Health Systems Research and Dynamical Modelling Unit Introduction to SEIR Models Nakul Chitnis Workshop on Mathematical Models of Climate Variability, Environmental
Department of Epidemiology and Public Health Health Systems Research and Dynamical Modelling Unit Introduction to SEIR Models Nakul Chitnis Workshop on Mathematical Models of Climate Variability, Environmental
THE x log x CONDITION FOR GENERAL BRANCHING PROCESSES
 J. Appl. Prob. 35, 537 544 (1998) Printed in Israel Applied Probability Trust 1998 THE x log x CONDITION FOR GENERAL BRANCHING PROCESSES PETER OLOFSSON, Rice University Abstract The x log x condition is
J. Appl. Prob. 35, 537 544 (1998) Printed in Israel Applied Probability Trust 1998 THE x log x CONDITION FOR GENERAL BRANCHING PROCESSES PETER OLOFSSON, Rice University Abstract The x log x condition is
MATH 56A: STOCHASTIC PROCESSES CHAPTER 2
 MATH 56A: STOCHASTIC PROCESSES CHAPTER 2 2. Countable Markov Chains I started Chapter 2 which talks about Markov chains with a countably infinite number of states. I did my favorite example which is on
MATH 56A: STOCHASTIC PROCESSES CHAPTER 2 2. Countable Markov Chains I started Chapter 2 which talks about Markov chains with a countably infinite number of states. I did my favorite example which is on
Sharpness of second moment criteria for branching and tree-indexed processes
 Sharpness of second moment criteria for branching and tree-indexed processes Robin Pemantle 1, 2 ABSTRACT: A class of branching processes in varying environments is exhibited which become extinct almost
Sharpness of second moment criteria for branching and tree-indexed processes Robin Pemantle 1, 2 ABSTRACT: A class of branching processes in varying environments is exhibited which become extinct almost
Hard-Core Model on Random Graphs
 Hard-Core Model on Random Graphs Antar Bandyopadhyay Theoretical Statistics and Mathematics Unit Seminar Theoretical Statistics and Mathematics Unit Indian Statistical Institute, New Delhi Centre New Delhi,
Hard-Core Model on Random Graphs Antar Bandyopadhyay Theoretical Statistics and Mathematics Unit Seminar Theoretical Statistics and Mathematics Unit Indian Statistical Institute, New Delhi Centre New Delhi,
1 Mechanistic and generative models of network structure
 1 Mechanistic and generative models of network structure There are many models of network structure, and these largely can be divided into two classes: mechanistic models and generative or probabilistic
1 Mechanistic and generative models of network structure There are many models of network structure, and these largely can be divided into two classes: mechanistic models and generative or probabilistic
Endemic persistence or disease extinction: the effect of separation into subcommunities
 Mathematical Statistics Stockholm University Endemic persistence or disease extinction: the effect of separation into subcommunities Mathias Lindholm Tom Britton Research Report 2006:6 ISSN 1650-0377 Postal
Mathematical Statistics Stockholm University Endemic persistence or disease extinction: the effect of separation into subcommunities Mathias Lindholm Tom Britton Research Report 2006:6 ISSN 1650-0377 Postal
Statistics 253/317 Introduction to Probability Models. Winter Midterm Exam Monday, Feb 10, 2014
 Statistics 253/317 Introduction to Probability Models Winter 2014 - Midterm Exam Monday, Feb 10, 2014 Student Name (print): (a) Do not sit directly next to another student. (b) This is a closed-book, closed-note
Statistics 253/317 Introduction to Probability Models Winter 2014 - Midterm Exam Monday, Feb 10, 2014 Student Name (print): (a) Do not sit directly next to another student. (b) This is a closed-book, closed-note
BRANCHING PROCESSES 1. GALTON-WATSON PROCESSES
 BRANCHING PROCESSES 1. GALTON-WATSON PROCESSES Galton-Watson processes were introduced by Francis Galton in 1889 as a simple mathematical model for the propagation of family names. They were reinvented
BRANCHING PROCESSES 1. GALTON-WATSON PROCESSES Galton-Watson processes were introduced by Francis Galton in 1889 as a simple mathematical model for the propagation of family names. They were reinvented
Modeling, Analysis, and Control of Information Propagation in Multi-layer and Multiplex Networks. Osman Yağan
 Modeling, Analysis, and Control of Information Propagation in Multi-layer and Multiplex Networks Osman Yağan Department of ECE Carnegie Mellon University Joint work with Y. Zhuang and V. Gligor (CMU) Alex
Modeling, Analysis, and Control of Information Propagation in Multi-layer and Multiplex Networks Osman Yağan Department of ECE Carnegie Mellon University Joint work with Y. Zhuang and V. Gligor (CMU) Alex
Age-dependent branching processes with incubation
 Age-dependent branching processes with incubation I. RAHIMOV Department of Mathematical Sciences, KFUPM, Box. 1339, Dhahran, 3161, Saudi Arabia e-mail: rahimov @kfupm.edu.sa We study a modification of
Age-dependent branching processes with incubation I. RAHIMOV Department of Mathematical Sciences, KFUPM, Box. 1339, Dhahran, 3161, Saudi Arabia e-mail: rahimov @kfupm.edu.sa We study a modification of
Random trees and branching processes
 Random trees and branching processes Svante Janson IMS Medallion Lecture 12 th Vilnius Conference and 2018 IMS Annual Meeting Vilnius, 5 July, 2018 Part I. Galton Watson trees Let ξ be a random variable
Random trees and branching processes Svante Janson IMS Medallion Lecture 12 th Vilnius Conference and 2018 IMS Annual Meeting Vilnius, 5 July, 2018 Part I. Galton Watson trees Let ξ be a random variable
Epidemics in Networks Part 2 Compartmental Disease Models
 Epidemics in Networks Part 2 Compartmental Disease Models Joel C. Miller & Tom Hladish 18 20 July 2018 1 / 35 Introduction to Compartmental Models Dynamics R 0 Epidemic Probability Epidemic size Review
Epidemics in Networks Part 2 Compartmental Disease Models Joel C. Miller & Tom Hladish 18 20 July 2018 1 / 35 Introduction to Compartmental Models Dynamics R 0 Epidemic Probability Epidemic size Review
14 Branching processes
 4 BRANCHING PROCESSES 6 4 Branching processes In this chapter we will consider a rom model for population growth in the absence of spatial or any other resource constraints. So, consider a population of
4 BRANCHING PROCESSES 6 4 Branching processes In this chapter we will consider a rom model for population growth in the absence of spatial or any other resource constraints. So, consider a population of
Transmission in finite populations
 Transmission in finite populations Juliet Pulliam, PhD Department of Biology and Emerging Pathogens Institute University of Florida and RAPIDD Program, DIEPS Fogarty International Center US National Institutes
Transmission in finite populations Juliet Pulliam, PhD Department of Biology and Emerging Pathogens Institute University of Florida and RAPIDD Program, DIEPS Fogarty International Center US National Institutes
Stochastic modelling of epidemic spread
 Stochastic modelling of epidemic spread Julien Arino Department of Mathematics University of Manitoba Winnipeg Julien Arino@umanitoba.ca 19 May 2012 1 Introduction 2 Stochastic processes 3 The SIS model
Stochastic modelling of epidemic spread Julien Arino Department of Mathematics University of Manitoba Winnipeg Julien Arino@umanitoba.ca 19 May 2012 1 Introduction 2 Stochastic processes 3 The SIS model
Network Modelling for Sexually. Transmitted Diseases
 Network Modelling for Sexually Transmitted Diseases Thitiya Theparod, MSc Bsc (Hons) Submitted for the degree of Doctor of Philosophy at Lancaster University 2015 Abstract The aim of this thesis is to
Network Modelling for Sexually Transmitted Diseases Thitiya Theparod, MSc Bsc (Hons) Submitted for the degree of Doctor of Philosophy at Lancaster University 2015 Abstract The aim of this thesis is to
Lecture 2: Tipping Point and Branching Processes
 ENAS-962 Theoretical Challenges in Networ Science Lecture 2: Tipping Point and Branching Processes Lecturer: Amin Karbasi Scribes: Amin Karbasi 1 The Tipping Point In most given systems, there is an inherent
ENAS-962 Theoretical Challenges in Networ Science Lecture 2: Tipping Point and Branching Processes Lecturer: Amin Karbasi Scribes: Amin Karbasi 1 The Tipping Point In most given systems, there is an inherent
Epidemics in Complex Networks and Phase Transitions
 Master M2 Sciences de la Matière ENS de Lyon 2015-2016 Phase Transitions and Critical Phenomena Epidemics in Complex Networks and Phase Transitions Jordan Cambe January 13, 2016 Abstract Spreading phenomena
Master M2 Sciences de la Matière ENS de Lyon 2015-2016 Phase Transitions and Critical Phenomena Epidemics in Complex Networks and Phase Transitions Jordan Cambe January 13, 2016 Abstract Spreading phenomena
Random graphs: Random geometric graphs
 Random graphs: Random geometric graphs Mathew Penrose (University of Bath) Oberwolfach workshop Stochastic analysis for Poisson point processes February 2013 athew Penrose (Bath), Oberwolfach February
Random graphs: Random geometric graphs Mathew Penrose (University of Bath) Oberwolfach workshop Stochastic analysis for Poisson point processes February 2013 athew Penrose (Bath), Oberwolfach February
A BINOMIAL MOMENT APPROXIMATION SCHEME FOR EPIDEMIC SPREADING IN NETWORKS
 U.P.B. Sci. Bull., Series A, Vol. 76, Iss. 2, 2014 ISSN 1223-7027 A BINOMIAL MOMENT APPROXIMATION SCHEME FOR EPIDEMIC SPREADING IN NETWORKS Yilun SHANG 1 Epidemiological network models study the spread
U.P.B. Sci. Bull., Series A, Vol. 76, Iss. 2, 2014 ISSN 1223-7027 A BINOMIAL MOMENT APPROXIMATION SCHEME FOR EPIDEMIC SPREADING IN NETWORKS Yilun SHANG 1 Epidemiological network models study the spread
UPPER DEVIATIONS FOR SPLIT TIMES OF BRANCHING PROCESSES
 Applied Probability Trust 7 May 22 UPPER DEVIATIONS FOR SPLIT TIMES OF BRANCHING PROCESSES HAMED AMINI, AND MARC LELARGE, ENS-INRIA Abstract Upper deviation results are obtained for the split time of a
Applied Probability Trust 7 May 22 UPPER DEVIATIONS FOR SPLIT TIMES OF BRANCHING PROCESSES HAMED AMINI, AND MARC LELARGE, ENS-INRIA Abstract Upper deviation results are obtained for the split time of a
Markov-modulated interactions in SIR epidemics
 Markov-modulated interactions in SIR epidemics E. Almaraz 1, A. Gómez-Corral 2 (1)Departamento de Estadística e Investigación Operativa, Facultad de Ciencias Matemáticas (UCM), (2)Instituto de Ciencias
Markov-modulated interactions in SIR epidemics E. Almaraz 1, A. Gómez-Corral 2 (1)Departamento de Estadística e Investigación Operativa, Facultad de Ciencias Matemáticas (UCM), (2)Instituto de Ciencias
MATHEMATICAL MODELS Vol. III - Mathematical Models in Epidemiology - M. G. Roberts, J. A. P. Heesterbeek
 MATHEMATICAL MODELS I EPIDEMIOLOGY M. G. Roberts Institute of Information and Mathematical Sciences, Massey University, Auckland, ew Zealand J. A. P. Heesterbeek Faculty of Veterinary Medicine, Utrecht
MATHEMATICAL MODELS I EPIDEMIOLOGY M. G. Roberts Institute of Information and Mathematical Sciences, Massey University, Auckland, ew Zealand J. A. P. Heesterbeek Faculty of Veterinary Medicine, Utrecht
Markov Chain Monte Carlo The Metropolis-Hastings Algorithm
 Markov Chain Monte Carlo The Metropolis-Hastings Algorithm Anthony Trubiano April 11th, 2018 1 Introduction Markov Chain Monte Carlo (MCMC) methods are a class of algorithms for sampling from a probability
Markov Chain Monte Carlo The Metropolis-Hastings Algorithm Anthony Trubiano April 11th, 2018 1 Introduction Markov Chain Monte Carlo (MCMC) methods are a class of algorithms for sampling from a probability
arxiv: v1 [math.pr] 13 Nov 2018
![arxiv: v1 [math.pr] 13 Nov 2018 arxiv: v1 [math.pr] 13 Nov 2018](/thumbs/90/102457202.jpg) PHASE TRANSITION FOR THE FROG MODEL ON BIREGULAR TREES ELCIO LEBENSZTAYN AND JAIME UTRIA arxiv:1811.05495v1 [math.pr] 13 Nov 2018 Abstract. We study the frog model with death on the biregular tree T d1,d
PHASE TRANSITION FOR THE FROG MODEL ON BIREGULAR TREES ELCIO LEBENSZTAYN AND JAIME UTRIA arxiv:1811.05495v1 [math.pr] 13 Nov 2018 Abstract. We study the frog model with death on the biregular tree T d1,d
Metric spaces and metrizability
 1 Motivation Metric spaces and metrizability By this point in the course, this section should not need much in the way of motivation. From the very beginning, we have talked about R n usual and how relatively
1 Motivation Metric spaces and metrizability By this point in the course, this section should not need much in the way of motivation. From the very beginning, we have talked about R n usual and how relatively
1/2 1/2 1/4 1/4 8 1/2 1/2 1/2 1/2 8 1/2 6 P =
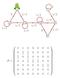 / 7 8 / / / /4 4 5 / /4 / 8 / 6 P = 0 0 0 0 0 0 0 0 0 0 4 0 0 0 0 0 0 0 0 4 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 Andrei Andreevich Markov (856 9) In Example. 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 P (n) = 0
/ 7 8 / / / /4 4 5 / /4 / 8 / 6 P = 0 0 0 0 0 0 0 0 0 0 4 0 0 0 0 0 0 0 0 4 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 Andrei Andreevich Markov (856 9) In Example. 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 P (n) = 0
Continuing the calculation of the absorption probability for a branching process. Homework 3 is due Tuesday, November 29.
 Extinction Probability for Branching Processes Friday, November 11, 2011 2:05 PM Continuing the calculation of the absorption probability for a branching process. Homework 3 is due Tuesday, November 29.
Extinction Probability for Branching Processes Friday, November 11, 2011 2:05 PM Continuing the calculation of the absorption probability for a branching process. Homework 3 is due Tuesday, November 29.
Problems on Evolutionary dynamics
 Problems on Evolutionary dynamics Doctoral Programme in Physics José A. Cuesta Lausanne, June 10 13, 2014 Replication 1. Consider the Galton-Watson process defined by the offspring distribution p 0 =
Problems on Evolutionary dynamics Doctoral Programme in Physics José A. Cuesta Lausanne, June 10 13, 2014 Replication 1. Consider the Galton-Watson process defined by the offspring distribution p 0 =
Susceptible-Infective-Removed Epidemics and Erdős-Rényi random
 Susceptible-Infective-Removed Epidemics and Erdős-Rényi random graphs MSR-Inria Joint Centre October 13, 2015 SIR epidemics: the Reed-Frost model Individuals i [n] when infected, attempt to infect all
Susceptible-Infective-Removed Epidemics and Erdős-Rényi random graphs MSR-Inria Joint Centre October 13, 2015 SIR epidemics: the Reed-Frost model Individuals i [n] when infected, attempt to infect all
Epidemics and information spreading
 Epidemics and information spreading Leonid E. Zhukov School of Data Analysis and Artificial Intelligence Department of Computer Science National Research University Higher School of Economics Social Network
Epidemics and information spreading Leonid E. Zhukov School of Data Analysis and Artificial Intelligence Department of Computer Science National Research University Higher School of Economics Social Network
Analytically tractable processes on networks
 University of California San Diego CERTH, 25 May 2011 Outline Motivation 1 Motivation Networks Random walk and Consensus Epidemic models Spreading processes on networks 2 Networks Motivation Networks Random
University of California San Diego CERTH, 25 May 2011 Outline Motivation 1 Motivation Networks Random walk and Consensus Epidemic models Spreading processes on networks 2 Networks Motivation Networks Random
 http://www.math.uah.edu/stat/markov/.xhtml 1 of 9 7/16/2009 7:20 AM Virtual Laboratories > 16. Markov Chains > 1 2 3 4 5 6 7 8 9 10 11 12 1. A Markov process is a random process in which the future is
http://www.math.uah.edu/stat/markov/.xhtml 1 of 9 7/16/2009 7:20 AM Virtual Laboratories > 16. Markov Chains > 1 2 3 4 5 6 7 8 9 10 11 12 1. A Markov process is a random process in which the future is
Any live cell with less than 2 live neighbours dies. Any live cell with 2 or 3 live neighbours lives on to the next step.
 2. Cellular automata, and the SIRS model In this Section we consider an important set of models used in computer simulations, which are called cellular automata (these are very similar to the so-called
2. Cellular automata, and the SIRS model In this Section we consider an important set of models used in computer simulations, which are called cellular automata (these are very similar to the so-called
Erdős-Renyi random graphs basics
 Erdős-Renyi random graphs basics Nathanaël Berestycki U.B.C. - class on percolation We take n vertices and a number p = p(n) with < p < 1. Let G(n, p(n)) be the graph such that there is an edge between
Erdős-Renyi random graphs basics Nathanaël Berestycki U.B.C. - class on percolation We take n vertices and a number p = p(n) with < p < 1. Let G(n, p(n)) be the graph such that there is an edge between
Latent voter model on random regular graphs
 Latent voter model on random regular graphs Shirshendu Chatterjee Cornell University (visiting Duke U.) Work in progress with Rick Durrett April 25, 2011 Outline Definition of voter model and duality with
Latent voter model on random regular graphs Shirshendu Chatterjee Cornell University (visiting Duke U.) Work in progress with Rick Durrett April 25, 2011 Outline Definition of voter model and duality with
Homework 6: Solutions Sid Banerjee Problem 1: (The Flajolet-Martin Counter) ORIE 4520: Stochastics at Scale Fall 2015
 Problem 1: (The Flajolet-Martin Counter) In class (and in the prelim!), we looked at an idealized algorithm for finding the number of distinct elements in a stream, where we sampled uniform random variables
Problem 1: (The Flajolet-Martin Counter) In class (and in the prelim!), we looked at an idealized algorithm for finding the number of distinct elements in a stream, where we sampled uniform random variables
CASE STUDY: EXTINCTION OF FAMILY NAMES
 CASE STUDY: EXTINCTION OF FAMILY NAMES The idea that families die out originated in antiquity, particilarly since the establishment of patrilineality (a common kinship system in which an individual s family
CASE STUDY: EXTINCTION OF FAMILY NAMES The idea that families die out originated in antiquity, particilarly since the establishment of patrilineality (a common kinship system in which an individual s family
Markov Chains CK eqns Classes Hitting times Rec./trans. Strong Markov Stat. distr. Reversibility * Markov Chains
 Markov Chains A random process X is a family {X t : t T } of random variables indexed by some set T. When T = {0, 1, 2,... } one speaks about a discrete-time process, for T = R or T = [0, ) one has a continuous-time
Markov Chains A random process X is a family {X t : t T } of random variables indexed by some set T. When T = {0, 1, 2,... } one speaks about a discrete-time process, for T = R or T = [0, ) one has a continuous-time
University of Leeds. Project in Statistics. Math5004M. Epidemic Modelling. Supervisor: Dr Andrew J Baczkowski. Student: Ross Breckon
 University of Leeds Project in Statistics Math5004M Epidemic Modelling Student: Ross Breckon 200602800 Supervisor: Dr Andrew J Baczkowski May 6, 2015 Abstract The outbreak of an infectious disease can
University of Leeds Project in Statistics Math5004M Epidemic Modelling Student: Ross Breckon 200602800 Supervisor: Dr Andrew J Baczkowski May 6, 2015 Abstract The outbreak of an infectious disease can
LIMITING PROBABILITY TRANSITION MATRIX OF A CONDENSED FIBONACCI TREE
 International Journal of Applied Mathematics Volume 31 No. 18, 41-49 ISSN: 1311-178 (printed version); ISSN: 1314-86 (on-line version) doi: http://dx.doi.org/1.173/ijam.v31i.6 LIMITING PROBABILITY TRANSITION
International Journal of Applied Mathematics Volume 31 No. 18, 41-49 ISSN: 1311-178 (printed version); ISSN: 1314-86 (on-line version) doi: http://dx.doi.org/1.173/ijam.v31i.6 LIMITING PROBABILITY TRANSITION
MCMC 2: Lecture 2 Coding and output. Phil O Neill Theo Kypraios School of Mathematical Sciences University of Nottingham
 MCMC 2: Lecture 2 Coding and output Phil O Neill Theo Kypraios School of Mathematical Sciences University of Nottingham Contents 1. General (Markov) epidemic model 2. Non-Markov epidemic model 3. Debugging
MCMC 2: Lecture 2 Coding and output Phil O Neill Theo Kypraios School of Mathematical Sciences University of Nottingham Contents 1. General (Markov) epidemic model 2. Non-Markov epidemic model 3. Debugging
LIMIT THEOREMS FOR A RANDOM GRAPH EPIDEMIC MODEL. By Håkan Andersson Stockholm University
 The Annals of Applied Probability 1998, Vol. 8, No. 4, 1331 1349 LIMIT THEOREMS FOR A RANDOM GRAPH EPIDEMIC MODEL By Håkan Andersson Stockholm University We consider a simple stochastic discrete-time epidemic
The Annals of Applied Probability 1998, Vol. 8, No. 4, 1331 1349 LIMIT THEOREMS FOR A RANDOM GRAPH EPIDEMIC MODEL By Håkan Andersson Stockholm University We consider a simple stochastic discrete-time epidemic
Lecture 4: September Reminder: convergence of sequences
 36-705: Intermediate Statistics Fall 2017 Lecturer: Siva Balakrishnan Lecture 4: September 6 In this lecture we discuss the convergence of random variables. At a high-level, our first few lectures focused
36-705: Intermediate Statistics Fall 2017 Lecturer: Siva Balakrishnan Lecture 4: September 6 In this lecture we discuss the convergence of random variables. At a high-level, our first few lectures focused
Stability theory is a fundamental topic in mathematics and engineering, that include every
 Stability Theory Stability theory is a fundamental topic in mathematics and engineering, that include every branches of control theory. For a control system, the least requirement is that the system is
Stability Theory Stability theory is a fundamental topic in mathematics and engineering, that include every branches of control theory. For a control system, the least requirement is that the system is
Mathematical Analysis of Epidemiological Models: Introduction
 Mathematical Analysis of Epidemiological Models: Introduction Jan Medlock Clemson University Department of Mathematical Sciences 8 February 2010 1. Introduction. The effectiveness of improved sanitation,
Mathematical Analysis of Epidemiological Models: Introduction Jan Medlock Clemson University Department of Mathematical Sciences 8 February 2010 1. Introduction. The effectiveness of improved sanitation,
HETEROGENEOUS MIXING IN EPIDEMIC MODELS
 CANADIAN APPLIED MATHEMATICS QUARTERLY Volume 2, Number 1, Spring 212 HETEROGENEOUS MIXING IN EPIDEMIC MODELS FRED BRAUER ABSTRACT. We extend the relation between the basic reproduction number and the
CANADIAN APPLIED MATHEMATICS QUARTERLY Volume 2, Number 1, Spring 212 HETEROGENEOUS MIXING IN EPIDEMIC MODELS FRED BRAUER ABSTRACT. We extend the relation between the basic reproduction number and the
Random Geometric Graphs
 Random Geometric Graphs Mathew D. Penrose University of Bath, UK Networks: stochastic models for populations and epidemics ICMS, Edinburgh September 2011 1 MODELS of RANDOM GRAPHS Erdos-Renyi G(n, p):
Random Geometric Graphs Mathew D. Penrose University of Bath, UK Networks: stochastic models for populations and epidemics ICMS, Edinburgh September 2011 1 MODELS of RANDOM GRAPHS Erdos-Renyi G(n, p):
Bootstrap Percolation on Periodic Trees
 Bootstrap Percolation on Periodic Trees Milan Bradonjić Iraj Saniee Abstract We study bootstrap percolation with the threshold parameter θ 2 and the initial probability p on infinite periodic trees that
Bootstrap Percolation on Periodic Trees Milan Bradonjić Iraj Saniee Abstract We study bootstrap percolation with the threshold parameter θ 2 and the initial probability p on infinite periodic trees that
Cover Page. The handle holds various files of this Leiden University dissertation
 Cover Page The handle http://hdl.handle.net/1887/39637 holds various files of this Leiden University dissertation Author: Smit, Laurens Title: Steady-state analysis of large scale systems : the successive
Cover Page The handle http://hdl.handle.net/1887/39637 holds various files of this Leiden University dissertation Author: Smit, Laurens Title: Steady-state analysis of large scale systems : the successive
Social Influence in Online Social Networks. Epidemiological Models. Epidemic Process
 Social Influence in Online Social Networks Toward Understanding Spatial Dependence on Epidemic Thresholds in Networks Dr. Zesheng Chen Viral marketing ( word-of-mouth ) Blog information cascading Rumor
Social Influence in Online Social Networks Toward Understanding Spatial Dependence on Epidemic Thresholds in Networks Dr. Zesheng Chen Viral marketing ( word-of-mouth ) Blog information cascading Rumor
CENOZOIC TECTONIC EVOLUTION OF THE IBERIAN PENINSULA: EFFECTS AND CAUSES OF CHANGING STRESS FIELDS
 VRIJE UNIVERSITEIT CENOZOIC TECTONIC EVOLUTION OF THE IBERIAN PENINSULA: EFFECTS AND CAUSES OF CHANGING STRESS FIELDS ACADEMISCH PROEFSCHRIFT ter verkrijging van de graad van doctor aan de Vrije Universiteit
VRIJE UNIVERSITEIT CENOZOIC TECTONIC EVOLUTION OF THE IBERIAN PENINSULA: EFFECTS AND CAUSES OF CHANGING STRESS FIELDS ACADEMISCH PROEFSCHRIFT ter verkrijging van de graad van doctor aan de Vrije Universiteit
Markov Chains. Arnoldo Frigessi Bernd Heidergott November 4, 2015
 Markov Chains Arnoldo Frigessi Bernd Heidergott November 4, 2015 1 Introduction Markov chains are stochastic models which play an important role in many applications in areas as diverse as biology, finance,
Markov Chains Arnoldo Frigessi Bernd Heidergott November 4, 2015 1 Introduction Markov chains are stochastic models which play an important role in many applications in areas as diverse as biology, finance,
Epidemic Models. Maaike Koninx July 12, Bachelor thesis Supervisor: dr. S.G. Cox
 Epidemic Models Maaike Koninx July 12, 2015 Bachelor thesis Supervisor: dr. S.G. Cox Korteweg-de Vries Instituut voor Wiskunde Faculteit der Natuurwetenschappen, Wiskunde en Informatica Universiteit van
Epidemic Models Maaike Koninx July 12, 2015 Bachelor thesis Supervisor: dr. S.G. Cox Korteweg-de Vries Instituut voor Wiskunde Faculteit der Natuurwetenschappen, Wiskunde en Informatica Universiteit van
CONSTRAINED PERCOLATION ON Z 2
 CONSTRAINED PERCOLATION ON Z 2 ZHONGYANG LI Abstract. We study a constrained percolation process on Z 2, and prove the almost sure nonexistence of infinite clusters and contours for a large class of probability
CONSTRAINED PERCOLATION ON Z 2 ZHONGYANG LI Abstract. We study a constrained percolation process on Z 2, and prove the almost sure nonexistence of infinite clusters and contours for a large class of probability
Linear-fractional branching processes with countably many types
 Branching processes and and their applications Badajoz, April 11-13, 2012 Serik Sagitov Chalmers University and University of Gothenburg Linear-fractional branching processes with countably many types
Branching processes and and their applications Badajoz, April 11-13, 2012 Serik Sagitov Chalmers University and University of Gothenburg Linear-fractional branching processes with countably many types
Explosion in branching processes and applications to epidemics in random graph models
 Explosion in branching processes and applications to epidemics in random graph models Júlia Komjáthy joint with Enrico Baroni and Remco van der Hofstad Eindhoven University of Technology Advances on Epidemics
Explosion in branching processes and applications to epidemics in random graph models Júlia Komjáthy joint with Enrico Baroni and Remco van der Hofstad Eindhoven University of Technology Advances on Epidemics
Lecture 06 01/31/ Proofs for emergence of giant component
 M375T/M396C: Topics in Complex Networks Spring 2013 Lecture 06 01/31/13 Lecturer: Ravi Srinivasan Scribe: Tianran Geng 6.1 Proofs for emergence of giant component We now sketch the main ideas underlying
M375T/M396C: Topics in Complex Networks Spring 2013 Lecture 06 01/31/13 Lecturer: Ravi Srinivasan Scribe: Tianran Geng 6.1 Proofs for emergence of giant component We now sketch the main ideas underlying
1 Informal definition of a C-M-J process
 (Very rough) 1 notes on C-M-J processes Andreas E. Kyprianou, Department of Mathematical Sciences, University of Bath, Claverton Down, Bath, BA2 7AY. C-M-J processes are short for Crump-Mode-Jagers processes
(Very rough) 1 notes on C-M-J processes Andreas E. Kyprianou, Department of Mathematical Sciences, University of Bath, Claverton Down, Bath, BA2 7AY. C-M-J processes are short for Crump-Mode-Jagers processes
Figure The Threshold Theorem of epidemiology
 K/a Figure 3 6. Assurne that K 1/ a < K 2 and K 2 / ß < K 1 (a) Show that the equilibrium solution N 1 =0, N 2 =0 of (*) is unstable. (b) Show that the equilibrium solutions N 2 =0 and N 1 =0, N 2 =K 2
K/a Figure 3 6. Assurne that K 1/ a < K 2 and K 2 / ß < K 1 (a) Show that the equilibrium solution N 1 =0, N 2 =0 of (*) is unstable. (b) Show that the equilibrium solutions N 2 =0 and N 1 =0, N 2 =K 2
A Natural Introduction to Probability Theory
 Ronald Meester A Natural Introduction to Probability Theory Second Edition Birkhäuser Basel Boston Berlin Author: Ronald Meester Faculteit der Exacte Wetenschappen Vrije Universiteit De Boelelaan 1081a
Ronald Meester A Natural Introduction to Probability Theory Second Edition Birkhäuser Basel Boston Berlin Author: Ronald Meester Faculteit der Exacte Wetenschappen Vrije Universiteit De Boelelaan 1081a
Statistical inference
 Statistical inference Contents 1. Main definitions 2. Estimation 3. Testing L. Trapani MSc Induction - Statistical inference 1 1 Introduction: definition and preliminary theory In this chapter, we shall
Statistical inference Contents 1. Main definitions 2. Estimation 3. Testing L. Trapani MSc Induction - Statistical inference 1 1 Introduction: definition and preliminary theory In this chapter, we shall
Citation for published version (APA): Hin, V. (2017). Ontogenesis: Eco-evolutionary perspective on life history complexity.
 UvA-DARE (Digital Academic Repository) Ontogenesis Hin, V. Link to publication Citation for published version (APA): Hin, V. (2017). Ontogenesis: Eco-evolutionary perspective on life history complexity.
UvA-DARE (Digital Academic Repository) Ontogenesis Hin, V. Link to publication Citation for published version (APA): Hin, V. (2017). Ontogenesis: Eco-evolutionary perspective on life history complexity.
Chris Bishop s PRML Ch. 8: Graphical Models
 Chris Bishop s PRML Ch. 8: Graphical Models January 24, 2008 Introduction Visualize the structure of a probabilistic model Design and motivate new models Insights into the model s properties, in particular
Chris Bishop s PRML Ch. 8: Graphical Models January 24, 2008 Introduction Visualize the structure of a probabilistic model Design and motivate new models Insights into the model s properties, in particular
Revisiting the Contact Process
 Revisiting the Contact Process Maria Eulália Vares Universidade Federal do Rio de Janeiro World Meeting for Women in Mathematics - July 31, 2018 The classical contact process G = (V,E) graph, locally finite.
Revisiting the Contact Process Maria Eulália Vares Universidade Federal do Rio de Janeiro World Meeting for Women in Mathematics - July 31, 2018 The classical contact process G = (V,E) graph, locally finite.
6.207/14.15: Networks Lecture 3: Erdös-Renyi graphs and Branching processes
 6.207/14.15: Networks Lecture 3: Erdös-Renyi graphs and Branching processes Daron Acemoglu and Asu Ozdaglar MIT September 16, 2009 1 Outline Erdös-Renyi random graph model Branching processes Phase transitions
6.207/14.15: Networks Lecture 3: Erdös-Renyi graphs and Branching processes Daron Acemoglu and Asu Ozdaglar MIT September 16, 2009 1 Outline Erdös-Renyi random graph model Branching processes Phase transitions
Hitting Probabilities
 Stat25B: Probability Theory (Spring 23) Lecture: 2 Hitting Probabilities Lecturer: James W. Pitman Scribe: Brian Milch 2. Hitting probabilities Consider a Markov chain with a countable state space S and
Stat25B: Probability Theory (Spring 23) Lecture: 2 Hitting Probabilities Lecturer: James W. Pitman Scribe: Brian Milch 2. Hitting probabilities Consider a Markov chain with a countable state space S and
Some Background Material
 Chapter 1 Some Background Material In the first chapter, we present a quick review of elementary - but important - material as a way of dipping our toes in the water. This chapter also introduces important
Chapter 1 Some Background Material In the first chapter, we present a quick review of elementary - but important - material as a way of dipping our toes in the water. This chapter also introduces important
Randomized Algorithms III Min Cut
 Chapter 11 Randomized Algorithms III Min Cut CS 57: Algorithms, Fall 01 October 1, 01 11.1 Min Cut 11.1.1 Problem Definition 11. Min cut 11..0.1 Min cut G = V, E): undirected graph, n vertices, m edges.
Chapter 11 Randomized Algorithms III Min Cut CS 57: Algorithms, Fall 01 October 1, 01 11.1 Min Cut 11.1.1 Problem Definition 11. Min cut 11..0.1 Min cut G = V, E): undirected graph, n vertices, m edges.
MATH 56A: STOCHASTIC PROCESSES CHAPTER 0
 MATH 56A: STOCHASTIC PROCESSES CHAPTER 0 0. Chapter 0 I reviewed basic properties of linear differential equations in one variable. I still need to do the theory for several variables. 0.1. linear differential
MATH 56A: STOCHASTIC PROCESSES CHAPTER 0 0. Chapter 0 I reviewed basic properties of linear differential equations in one variable. I still need to do the theory for several variables. 0.1. linear differential
Discrete random structures whose limits are described by a PDE: 3 open problems
 Discrete random structures whose limits are described by a PDE: 3 open problems David Aldous April 3, 2014 Much of my research has involved study of n limits of size n random structures. There are many
Discrete random structures whose limits are described by a PDE: 3 open problems David Aldous April 3, 2014 Much of my research has involved study of n limits of size n random structures. There are many
Lecture 5: The Principle of Deferred Decisions. Chernoff Bounds
 Randomized Algorithms Lecture 5: The Principle of Deferred Decisions. Chernoff Bounds Sotiris Nikoletseas Associate Professor CEID - ETY Course 2013-2014 Sotiris Nikoletseas, Associate Professor Randomized
Randomized Algorithms Lecture 5: The Principle of Deferred Decisions. Chernoff Bounds Sotiris Nikoletseas Associate Professor CEID - ETY Course 2013-2014 Sotiris Nikoletseas, Associate Professor Randomized
Shortest Paths in Random Intersection Graphs
 Shortest Paths in Random Intersection Graphs Gesine Reinert Department of Statistics University of Oxford reinert@stats.ox.ac.uk September 14 th, 2011 1 / 29 Outline Bipartite graphs and random intersection
Shortest Paths in Random Intersection Graphs Gesine Reinert Department of Statistics University of Oxford reinert@stats.ox.ac.uk September 14 th, 2011 1 / 29 Outline Bipartite graphs and random intersection
Markov Chains and Pandemics
 Markov Chains and Pandemics Caleb Dedmore and Brad Smith December 8, 2016 Page 1 of 16 Abstract Markov Chain Theory is a powerful tool used in statistical analysis to make predictions about future events
Markov Chains and Pandemics Caleb Dedmore and Brad Smith December 8, 2016 Page 1 of 16 Abstract Markov Chain Theory is a powerful tool used in statistical analysis to make predictions about future events
UvA-DARE (Digital Academic Repository) Phenotypic variation in plants Lauss, K. Link to publication
 UvA-DARE (Digital Academic Repository) Phenotypic variation in plants Lauss, K. Link to publication Citation for published version (APA): Lauss, K. (2017). Phenotypic variation in plants: Roles for epigenetics
UvA-DARE (Digital Academic Repository) Phenotypic variation in plants Lauss, K. Link to publication Citation for published version (APA): Lauss, K. (2017). Phenotypic variation in plants: Roles for epigenetics
Connectedness. Proposition 2.2. The following are equivalent for a topological space (X, T ).
 Connectedness 1 Motivation Connectedness is the sort of topological property that students love. Its definition is intuitive and easy to understand, and it is a powerful tool in proofs of well-known results.
Connectedness 1 Motivation Connectedness is the sort of topological property that students love. Its definition is intuitive and easy to understand, and it is a powerful tool in proofs of well-known results.
Example: physical systems. If the state space. Example: speech recognition. Context can be. Example: epidemics. Suppose each infected
 4. Markov Chains A discrete time process {X n,n = 0,1,2,...} with discrete state space X n {0,1,2,...} is a Markov chain if it has the Markov property: P[X n+1 =j X n =i,x n 1 =i n 1,...,X 0 =i 0 ] = P[X
4. Markov Chains A discrete time process {X n,n = 0,1,2,...} with discrete state space X n {0,1,2,...} is a Markov chain if it has the Markov property: P[X n+1 =j X n =i,x n 1 =i n 1,...,X 0 =i 0 ] = P[X
Solutions For Stochastic Process Final Exam
 Solutions For Stochastic Process Final Exam (a) λ BMW = 20 0% = 2 X BMW Poisson(2) Let N t be the number of BMWs which have passes during [0, t] Then the probability in question is P (N ) = P (N = 0) =
Solutions For Stochastic Process Final Exam (a) λ BMW = 20 0% = 2 X BMW Poisson(2) Let N t be the number of BMWs which have passes during [0, t] Then the probability in question is P (N ) = P (N = 0) =
