Count Predicted Variable & Contingency Tables
|
|
|
- Hugo Chandler
- 6 years ago
- Views:
Transcription
1 Count Predicted Variable & Contingency Tables Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information.
2 Goals and General Idea
3 Goals Contingency tables When we have count data distributed across a range of different categories Are counts in one category higher than another? Is the count in one category contingent upon the the level in another category (or plural)? Eye Colour Hair Colour Blue Brown Green Hazel Black Blond Brunette Red Data from Snee (1974) The American Statistician 28: 9-12, as presented in Kruschke (2015).
4 Goals Contingency tables When we have count data distributed across a range of different categories Are counts in one category higher than another? Is the count in one category contingent upon the the level in another category (or plural)? Often addressed with chi-square or Exact test analyses
5 Goals Count predicted variable Any time our predicted variable is a count Abundance Counts of individuals (or things) with different traits/characteristics etc.
6 Distribution and Links When modelling count data, it is appropriate to use the Poisson distribution Positive integers One parameter - lambda (λ) λ = 1 λ = 3 Density Density X Values X Values λ = 5 λ = 10 Density Density X Values X Values
7 Distribution and Links Contingency table example Cell frequencies are representative of underlying cell probabilities Are nominal variables independent of each other?
8 Distribution and Links Contingency table example Cell frequencies are representative of underlying cell probabilities Are nominal variables independent of each other? If independent 68 = Pr(BlackHair) x Pr(BrownEyes) True for all cells
9 Distribution and Links Contingency table example Cell frequencies are representative of underlying cell probabilities Are nominal variables independent of each other? If independent 68 = Pr(BlackHair) x Pr(BrownEyes) True for all cells If interaction effects, this will not be the case
10 Distribution and Links Contingency table example Eye Colour Hair Colour Blue Brown Green Hazel Black Blond Brunette Red f(c) N marginal frequencies of each eye colour
11 Distribution and Links Contingency table example Eye Colour Hair Colour Blue Brown Green Hazel Black Blond Brunette Red f(c) N marginal frequencies of each eye colour Pr(BlueEyes) = 215 / 592 = 0.363
12 Distribution and Links Contingency table example Eye Colour Hair Colour Blue Brown Green Hazel Black Blond Brunette Red f(c) N marginal frequencies of each eye colour marginal frequencies of each hair colour Pr(BlueEyes) = 215 / 592 = 0.363
13 Distribution and Links Contingency table example Eye Colour Hair Colour Blue Brown Green Hazel Black Blond Brunette Red f(c) N marginal frequencies of each eye colour marginal frequencies of each hair colour Pr(BlueEyes) = 215 / 592 = Pr(BlackHair) = 108 / 592 = 0.182
14 Distribution and Links Contingency table example Eye Colour Hair Colour Blue Brown Green Hazel Black Blond Brunette Red f(c) N marginal frequencies of each eye colour marginal frequencies of each hair colour Pr(BlueEyes) = 215 / 592 = Pr(BlackHair) = 108 / 592 = Pr(BlueEyes & BlackHair) = (0.363 x 0.182) x 592 = 39 Hmm...must be an interaction effect
15 Distribution and Links Contingency table example Pr(BlueEyes & BlackHair) = (0.363 x 0.182) x 592 = 39 Joint probability is the product of the relevant marginal probabilities We re used to dealing with additive combinations Can convert to log scale, then they ll be additive
16 Segue on Logarithms Adding logarithms is the same as multiplying original values 10 x 5 = 50 log(10) + log(5) = log(50)
17 Segue on Logarithms Adding logarithms is the same as multiplying original values 10 x 5 = 50 log(10) + log(5) = log(50) Can cancel out logarithms (bring back to original scale), by raising them to the exponent exp(log(10) + log(5)) = 50
18 Distribution and Links
19 Distribution and Links The black box into which we can put any of our previous equations, or others
20 Distribution and Links Depends on rest of model, here the average across all categories of all variables
21 Distribution and Links The deflection away from baseline due to being in each category of our first nominal predictor variable.
22 Distribution and Links The deflection away from baseline due to being in each category of our second nominal predictor variable.
23 Distribution and Links Interaction effects. Must also be constrained so that they sum to zero
24 Distribution and Links Note that coefficient estimates will now be on the log scale (even though we haven t explicitly specified them as such)
25 Distribution and Links The link between the predictor and predicted variables is based on logarithms Previous models (other than logistic) have been based on identity These types of models are called log linear models
26 Frequentist Approach
27 Frequentist Approach Read data into R haireye <- read.table( haireye.csv, header = TRUE, sep =, )
28 Frequentist Approach First, organize the data y <- haireye$freq eye <- haireye$eye hair <- haireye$hair
29 Frequentist Approach Use the glm function (generalized linear model) Set family argument to poisson(link = log ) glm.out <- glm(y ~ eye + hair + (eye * hair), family = poisson(link = "log"))
30 Frequentist Approach summary(glm.out) Coefficients: Estimate Std. Error z value Pr(> z ) (Intercept) < 2e-16 *** eyebrown e-06 *** eyegreen ** eyehazel hairblond e-10 *** hairbrunette e-09 *** hairred eyebrown:hairblond e-16 *** eyegreen:hairblond eyehazel:hairblond e-05 *** eyebrown:hairbrunette ** eyegreen:hairbrunette eyehazel:hairbrunette eyebrown:hairred * eyegreen:hairred eyehazel:hairred
31 Frequentist Approach summary(glm.out) Coefficients: Estimate Std. Error z value Pr(> z ) (Intercept) < 2e-16 *** eyebrown e-06 *** eyegreen ** eyehazel hairblond e-10 *** hairbrunette Combined effect of blue eyes 8.03e-09 & black *** hairred hair (independently, and in all eyebrown:hairblond e-16 *** interactions) eyegreen:hairblond eyehazel:hairblond e-05 *** eyebrown:hairbrunette ** eyegreen:hairbrunette eyehazel:hairbrunette eyebrown:hairred * eyegreen:hairred eyehazel:hairred
32 Bayesian Approach
33 Load Libraries & Functions library(runjags) library(coda) source("plotpost.r")
34 Organize the Data # Y-Data y <- haireye$freq N <- length(y) ylogmean <- log(mean(y)) ylogsd <- log(sd(c(rep(0, N - 1), sum(y)))) # Eye data x1 <- as.numeric(haireye$eye) eyecolours <- levels(haireye$eye) neyecolour <- length(unique(x1)) # Hair data x2 <- as.numeric(haireye$hair) haircolours <- levels(haireye$hair) nhaircolour <- length(unique(x2))
35 Organize the Data # Y-Data y <- haireye$freq N <- length(y) ylogmean <- log(mean(y)) ylogsd <- log(sd(c(rep(0, N - 1), sum(y)))) # Eye data x1 <- as.numeric(haireye$eye) eyecolours <- levels(haireye$eye) neyecolour <- length(unique(x1)) # Hair data x2 <- as.numeric(haireye$hair) haircolours <- levels(haireye$hair) nhaircolour <- length(unique(x2)) Will see why we need these in a minute...
36 Organize the Data # Y-Data y <- haireye$freq N <- length(y) ylogmean <- log(mean(y)) ylogsd <- log(sd(c(rep(0, N - 1), sum(y)))) # Eye data x1 <- as.numeric(haireye$eye) eyecolours <- levels(haireye$eye) neyecolour <- length(unique(x1)) # Hair data x2 <- as.numeric(haireye$hair) haircolours <- levels(haireye$hair) nhaircolour <- length(unique(x2)) Largest possible sd would occur if the sum of all values was in one cell, and all others were zero
37 Make Data List For JAGS datalist = list( y = y, N = N, ylogmean = ylogmean, ylogsd = ylogsd, x1 = x1, x2 = x2, neyecolours = neyecolours, nhaircolours = nhaircolours )
38 Define the Model Model the same as what we ve encountered, except y values based on a Poisson distribution Link between predictors and predicted is the exponent
39 Define the Model 0 Poisson λi yi
40 Define the Model ylogmean (2 * ylogsd) µ τ = 1/σ 2 - norm λi 0 Poisson yi
41 Define the Model ylogmean (2 * ylogsd) α gamma β µ τ = 1/σ 2 - norm µ τ = 1/σ 2 norm 0 Poisson λi yi
42 Define the Model modelstring = " model { # The likelihood for (i in 1:N) { y[i] ~ dpois(lambda[i]) lambda[i] <- exp(a0 + a1[x1[i]] + a2[x2[i]] + a1a2[x1[i], x2[i]]) }...
43 Define the Model modelstring = " model { a instead of b because we still have to modify them so that the sum to zero within each variable (they are nominal) # The likelihood for (i in 1:N) { y[i] ~ dpois(lambda[i]) lambda[i] <- exp(a0 + a1[x1[i]] + a2[x2[i]] + a1a2[x1[i], x2[i]]) }...
44 Define the Model... # The Priors # a0 a0 ~ dnorm(ylogmean, 1 / ((2 * ylogsd)^2)) # a1 for (j in 1:nEyeColours) { a1[j] ~ dnorm(0, 1 / sigma1[j]^2) sigma1[j] ~ dgamma(1.1, 0.11) } # a2 for (j in 1:nHairColours) { a2[j] ~ dnorm(0, 1 / sigma2[j]^2) sigma2[j] ~ dgamma(1.1, 0.11) }... # a1a2 for (j in 1:nHairColours) { for (k in 1:nEyeColours) { a1a2[j, k] ~ dnorm(0, 1 / sigma12[j, k]^2) sigma12[j, k] ~ dgamma(1.1, 0.11) } }
45 Define the Model... # Convert variables to sum-to-zero for (j in 1:nEyeColours) { for (k in 1:nHairColours) { m[j, k] <- a0 + a1[j] + a2[k] + a1a2[j, k] # Cell means } } b0 <- mean(m[1:neyecolours, 1:nHairColours]) for (j in 1:nEyeColours) { b1[j] <- mean(m[j, 1:nHairColours]) - b0 } for (j in 1:nHairColours) { b2[j] <- mean(m[1:neyecolours, j]) - b0 } for (j in 1:nEyeColours) { for (k in 1:nHairColours) { b1b2[j, k] <- m[j, k] - (b0 + b1[j] + b2[k]) } } } " writelines(modelstring, con = "model.txt")
46 Specify Initial Values initslist <- function() { list( a0 = rnorm(n = 1, mean = ylogmean, sd = ylogsd), a1 = rnorm(n = neyecolours, mean = 0, sd = 10), a2 = rnorm(n = nhaircolours, mean = 0, sd = 10), sigma1 = rgamma(n = neyecolours, shape = 1.1, rate = 0.11), sigma2 = rgamma(n = nhaircolours, shape = 1.1, rate = 0.11) ) }
47 Specify Initial Values initslist <- function() { list( a0 = rnorm(n = 1, mean = ylogmean, sd = ylogsd), a1 = rnorm(n = neyecolours, mean = 0, sd = 10), a2 = rnorm(n = nhaircolours, mean = 0, sd = 10), sigma1 = rgamma(n = neyecolours, shape = 1.1, rate = 0.11), sigma2 = rgamma(n = nhaircolours, shape = 1.1, rate = 0.11) ) } Note absence of initial values for a1a2 and sigma12
48 Specify MCMC Parameters and Run runjagsout <- run.jags( method = "simple", model = "model.txt", monitor = c("b0", "b1", "b2", b1b2, sigma1, sigma2, sigma12 ), data = datalist, inits = initslist, n.chains = 3, adapt = 500, burnin = 1000, sample = 20000, thin = 1, summarise = TRUE, plots = FALSE)
49 Next Steps (On Your Own) Retrieve the data and take a peek at the structure Test model performance Extract & parse results
50 View Posteriors
51 Plotting Posterior Distributions β 0 par(mfrow = c(1, 1)) histinfo = plotpost(b0, xlab = bquote(beta[0])) mean = % HDI β 0
52 Plotting Posterior Distributions β 1 par(mfrow = c(2, 2)) for (i in 1:nEyeColours) { histinfo = plotpost(b1[i, ], xlab = bquote(beta * 1[.(i)]), main = paste("b1:", eyecolours[i])) }
53 Plotting Posterior Distributions β 1 b1: Blue b1: Brown mean = mean = % HDI % HDI β β1 2 b1: Green b1: Hazel mean = mean = % HDI % HDI β β1 4
54 Plotting Posterior Distributions β 1 b1: Blue b1: Brown mean = mean = Blue & brown eyes more frequent, green & hazel less so 95% HDI β1 1 95% HDI β1 2 b1: Green b1: Hazel mean = mean = % HDI % HDI β β1 4
55 Plotting Posterior Distributions β 2 par(mfrow = c(2, 2)) for (i in 1:nHairColours) { histinfo = plotpost(b2[i, ], xlab = bquote(beta * 2[.(i)]), main = paste("b2:", haircolours[i])) }
56 Plotting Posterior Distributions β 2 b2: Black b2: Blond mean = mean = Brunettes more frequent, all others less so 95% HDI β2 1 95% HDI β2 2 b2: Brunette b2: Red mean = mean = % HDI % HDI β β2 4
57 Plotting Posterior Distributions β 1 β 2 par(mfrow = c(2, 2)) ncombos = neyecolours * nhaircolours counter = 1 while (counter <= ncombos) { for (i in 1:nEyeColours) { for (j in 1:nHairColours) { histinfo = plotpost(b1b2[counter, ], xlab = bquote(beta * 1[.(i)] * beta * 2[.(j)]), main = paste("b1:", eyecolours[i], ", b2:", haircolours[j])) counter = counter + 1 } } }
58 Plotting Posterior Distributions β 1 β 2 b1: Blue, b2: Black b1: Blue, b2: Blond mean = mean = % HDI % HDI β1 1 β β1 1 β2 2 b1: Blue, b2: Brunette b1: Blue, b2: Red mean = mean = % HDI % HDI β1 1 β β1 1 β2 4
59 Plotting Posterior Distributions β 1 β 2 Blue eyes & blonde hair found together at a higher frequency than expected if no interaction b1: Blue, b2: Black mean = b1: Blue, b2: Blond mean = % HDI % HDI β1 1 β β1 1 β2 2 b1: Blue, b2: Brunette mean = b1: Blue, b2: Red mean = % HDI % HDI β1 1 β β1 1 β2 4
60 Plotting Posterior Distributions β 1 β 2 b1: Blue, b2: Black mean = Blue eyes & other combinations found together less often than expected b1: Blue, b2: Blond mean = % HDI % HDI β1 1 β β1 1 β2 2 b1: Blue, b2: Brunette b1: Blue, b2: Red mean = mean = % HDI % HDI β1 1 β β1 1 β2 4
61 Plotting Posterior Distributions β 1 β 2 b1: Green, b2: Black b1: Green, b2: Blond mean = mean = % HDI % HDI β1 3 β β1 3 β2 2 b1: Green, b2: Brunette b1: Green, b2: Red mean = mean = % HDI % HDI β1 3 β β1 3 β2 4
62 Plotting Posterior Distributions β 1 β 2 b1: Brown, b2: Black b1: Brown, b2: Blond mean = mean = % HDI % HDI β1 2 β β1 2 β2 2 b1: Brown, b2: Brunette b1: Brown, b2: Red mean = mean = % HDI % HDI β1 2 β β1 2 β2 4
63 Plotting Posterior Distributions β 1 β 2 b1: Hazel, b2: Black b1: Hazel, b2: Blond mean = mean = % HDI % HDI β1 4 β β1 4 β2 2 b1: Hazel, b2: Brunette b1: Hazel, b2: Red mean = mean = % HDI % HDI β1 4 β β1 4 β2 4
64 Calculating Expected Values
65 Calculating Expected Values Same as before with multiple nominal predictors Expected frequency of those with blue eyes & black hair par(mfrow = c(1, 1)) expblueblack <- exp(b0 + b1[1, ] + b2[1, ] + b1b2[1, ]) histinfo = plotpost(expblueblack, xlab = "Frequency", main = "Exp. Blue Eyes Black Hair")
66 Calculating Expected Values Same as before with multiple nominal predictors Row in b1 representing blue eyes Expected frequency of those with blue eyes & black hair par(mfrow = c(1, 1)) expblueblack <- exp(b0 + b1[1, ] + b2[1, ] + b1b2[1, ]) histinfo = plotpost(expblueblack, xlab = "Frequency", main = "Exp. Blue Eyes Black Hair")
67 Calculating Expected Values Same as before with multiple nominal predictors Row in b2 representing black hair Expected frequency of those with blue eyes & black hair par(mfrow = c(1, 1)) expblueblack <- exp(b0 + b1[1, ] + b2[1, ] + b1b2[1, ]) histinfo = plotpost(expblueblack, xlab = "Frequency", main = "Exp. Blue Eyes Black Hair")
68 Calculating Expected Values Row in b1b2 representing blue eyes & black hair interaction Same as before with multiple nominal predictors Expected frequency of those with blue eyes & black hair par(mfrow = c(1, 1)) expblueblack <- exp(b0 + b1[1, ] + b2[1, ] + b1b2[1, ]) histinfo = plotpost(expblueblack, xlab = "Frequency", main = "Exp. Blue Eyes Black Hair")
69 Calculating Expected Values Exp. Blue Eyes Black Hair mean = % HDI Frequency
70 Calculating Expected Values Expected frequency of those with blue eyes & black hair expgreenblond <- exp(b0 + b1[3, ] + b2[2, ] + b1b2[10, ]) histinfo = plotpost(expgreenblond, xlab = "Frequency", main = "Exp. Green Eyes Blond Hair")
71 Calculating Expected Values Exp. Green Eyes Blond Hair mean = % HDI Frequency
72 Posterior Predictive Check
73 Posterior Predictive Check Same as before with multiple nominal predictors for (i in 1:nPred) { for (j in 1:postSampSize) { ypostpred[i, j] <- rpois(n = 1, lambda = exp(b0[j] + b1[x1[i], j] + b2[x2[i], j] + b1b2[((x1[i] * 4) - 3) + (x2[i] - 1), j])) } }
74 Posterior Predictive Check Same as before with multiple nominal predictors for (i in 1:nPred) { for (j in 1:postSampSize) { ypostpred[i, j] <- rpois(n = 1, lambda = exp(b0[j] + b1[x1[i], j] + b2[x2[i], j] + b1b2[((x1[i] * 4) - 3) + (x2[i] - 1), j])) } } Using the Poisson distribution
75 Posterior Predictive Check Same as before with multiple nominal predictors for (i in 1:nPred) { for (j in 1:postSampSize) { ypostpred[i, j] <- rpois(n = 1, lambda = exp(b0[j] + b1[x1[i], j] + b2[x2[i], j] + b1b2[((x1[i] * 4) - 3) + (x2[i] - 1), j])) } } Raising the predictors to the exponent
76 Posterior Predictive Check Same as before with multiple nominal predictors for (i in 1:nPred) { for (j in 1:postSampSize) { ypostpred[i, j] <- rpois(n = 1, lambda = exp(b0[j] + b1[x1[i], j] + b2[x2[i], j] + b1b2[((x1[i] * 4) - 3) + (x2[i] - 1), j])) } } Have to figure out how to identify proper interaction coefficient row based on values in each predictor
77 Posterior Predictive Check Not bad! Frequency
78 Other Things
79 Other Things Can do all the things we did with other nominal data sets Contrasts between individual groups Contrasts between one group and all others Contrasts between any combination
80 Questions?
81 Creative Commons License Anyone is allowed to distribute, remix, tweak, and build upon this work, even commercially, as long as they credit me for the original creation. See the Creative Commons website for more information. Click here to go back to beginning
Multiple Regression: Nominal Predictors. Tim Frasier
 Multiple Regression: Nominal Predictors Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Goals
Multiple Regression: Nominal Predictors Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Goals
Metric Predicted Variable With One Nominal Predictor Variable
 Metric Predicted Variable With One Nominal Predictor Variable Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more
Metric Predicted Variable With One Nominal Predictor Variable Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more
Multiple Regression: Mixed Predictor Types. Tim Frasier
 Multiple Regression: Mixed Predictor Types Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. The
Multiple Regression: Mixed Predictor Types Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. The
Hierarchical Modeling
 Hierarchical Modeling Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. General Idea One benefit
Hierarchical Modeling Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. General Idea One benefit
Metric Predicted Variable on One Group
 Metric Predicted Variable on One Group Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Prior Homework
Metric Predicted Variable on One Group Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Prior Homework
Metric Predicted Variable on Two Groups
 Metric Predicted Variable on Two Groups Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Goals
Metric Predicted Variable on Two Groups Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Goals
Bayesian Statistics: An Introduction
 : An Introduction Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Outline 1. Bayesian statistics,
: An Introduction Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Outline 1. Bayesian statistics,
Introduction to R, Part I
 Introduction to R, Part I Basic math, variables, and variable types Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here
Introduction to R, Part I Basic math, variables, and variable types Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here
Linear Regression. Data Model. β, σ 2. Process Model. ,V β. ,s 2. s 1. Parameter Model
 Regression: Part II Linear Regression y~n X, 2 X Y Data Model β, σ 2 Process Model Β 0,V β s 1,s 2 Parameter Model Assumptions of Linear Model Homoskedasticity No error in X variables Error in Y variables
Regression: Part II Linear Regression y~n X, 2 X Y Data Model β, σ 2 Process Model Β 0,V β s 1,s 2 Parameter Model Assumptions of Linear Model Homoskedasticity No error in X variables Error in Y variables
Bayesian Graphical Models
 Graphical Models and Inference, Lecture 16, Michaelmas Term 2009 December 4, 2009 Parameter θ, data X = x, likelihood L(θ x) p(x θ). Express knowledge about θ through prior distribution π on θ. Inference
Graphical Models and Inference, Lecture 16, Michaelmas Term 2009 December 4, 2009 Parameter θ, data X = x, likelihood L(θ x) p(x θ). Express knowledge about θ through prior distribution π on θ. Inference
36-463/663: Multilevel & Hierarchical Models HW09 Solution
 36-463/663: Multilevel & Hierarchical Models HW09 Solution November 15, 2016 Quesion 1 Following the derivation given in class, when { n( x µ) 2 L(µ) exp, f(p) exp 2σ 2 0 ( the posterior is also normally
36-463/663: Multilevel & Hierarchical Models HW09 Solution November 15, 2016 Quesion 1 Following the derivation given in class, when { n( x µ) 2 L(µ) exp, f(p) exp 2σ 2 0 ( the posterior is also normally
Generalized Linear Models
 Generalized Linear Models Assumptions of Linear Model Homoskedasticity Model variance No error in X variables Errors in variables No missing data Missing data model Normally distributed error Error in
Generalized Linear Models Assumptions of Linear Model Homoskedasticity Model variance No error in X variables Errors in variables No missing data Missing data model Normally distributed error Error in
STAT Lecture 11: Bayesian Regression
 STAT 491 - Lecture 11: Bayesian Regression Generalized Linear Models Generalized linear models (GLMs) are a class of techniques that include linear regression, logistic regression, and Poisson regression.
STAT 491 - Lecture 11: Bayesian Regression Generalized Linear Models Generalized linear models (GLMs) are a class of techniques that include linear regression, logistic regression, and Poisson regression.
Normal distribution We have a random sample from N(m, υ). The sample mean is Ȳ and the corrected sum of squares is S yy. After some simplification,
 Likelihood Let P (D H) be the probability an experiment produces data D, given hypothesis H. Usually H is regarded as fixed and D variable. Before the experiment, the data D are unknown, and the probability
Likelihood Let P (D H) be the probability an experiment produces data D, given hypothesis H. Usually H is regarded as fixed and D variable. Before the experiment, the data D are unknown, and the probability
Two Hours. Mathematical formula books and statistical tables are to be provided THE UNIVERSITY OF MANCHESTER. 26 May :00 16:00
 Two Hours MATH38052 Mathematical formula books and statistical tables are to be provided THE UNIVERSITY OF MANCHESTER GENERALISED LINEAR MODELS 26 May 2016 14:00 16:00 Answer ALL TWO questions in Section
Two Hours MATH38052 Mathematical formula books and statistical tables are to be provided THE UNIVERSITY OF MANCHESTER GENERALISED LINEAR MODELS 26 May 2016 14:00 16:00 Answer ALL TWO questions in Section
Generalized Linear Models
 Generalized Linear Models Assumptions of Linear Model Homoskedasticity Model variance No error in X variables Errors in variables No missing data Missing data model Normally distributed error GLM Error
Generalized Linear Models Assumptions of Linear Model Homoskedasticity Model variance No error in X variables Errors in variables No missing data Missing data model Normally distributed error GLM Error
Statistics in Environmental Research (BUC Workshop Series) II Problem sheet - WinBUGS - SOLUTIONS
 Statistics in Environmental Research (BUC Workshop Series) II Problem sheet - WinBUGS - SOLUTIONS 1. (a) The posterior mean estimate of α is 14.27, and the posterior mean for the standard deviation of
Statistics in Environmental Research (BUC Workshop Series) II Problem sheet - WinBUGS - SOLUTIONS 1. (a) The posterior mean estimate of α is 14.27, and the posterior mean for the standard deviation of
Robust Bayesian Regression
 Readings: Hoff Chapter 9, West JRSSB 1984, Fúquene, Pérez & Pericchi 2015 Duke University November 17, 2016 Body Fat Data: Intervals w/ All Data Response % Body Fat and Predictor Waist Circumference 95%
Readings: Hoff Chapter 9, West JRSSB 1984, Fúquene, Pérez & Pericchi 2015 Duke University November 17, 2016 Body Fat Data: Intervals w/ All Data Response % Body Fat and Predictor Waist Circumference 95%
WinBUGS : part 2. Bruno Boulanger Jonathan Jaeger Astrid Jullion Philippe Lambert. Gabriele, living with rheumatoid arthritis
 WinBUGS : part 2 Bruno Boulanger Jonathan Jaeger Astrid Jullion Philippe Lambert Gabriele, living with rheumatoid arthritis Agenda 2! Hierarchical model: linear regression example! R2WinBUGS Linear Regression
WinBUGS : part 2 Bruno Boulanger Jonathan Jaeger Astrid Jullion Philippe Lambert Gabriele, living with rheumatoid arthritis Agenda 2! Hierarchical model: linear regression example! R2WinBUGS Linear Regression
Generalized Linear Models
 Generalized Linear Models STAT 489-01: Bayesian Methods of Data Analysis Spring Semester 2017 Contents 1 Multi-Variable Linear Models 1 2 Generalized Linear Models 4 2.1 Illustration: Logistic Regression..........
Generalized Linear Models STAT 489-01: Bayesian Methods of Data Analysis Spring Semester 2017 Contents 1 Multi-Variable Linear Models 1 2 Generalized Linear Models 4 2.1 Illustration: Logistic Regression..........
Bayesian Inference for Regression Parameters
 Bayesian Inference for Regression Parameters 1 Bayesian inference for simple linear regression parameters follows the usual pattern for all Bayesian analyses: 1. Form a prior distribution over all unknown
Bayesian Inference for Regression Parameters 1 Bayesian inference for simple linear regression parameters follows the usual pattern for all Bayesian analyses: 1. Form a prior distribution over all unknown
36-463/663Multilevel and Hierarchical Models
 36-463/663Multilevel and Hierarchical Models From Bayes to MCMC to MLMs Brian Junker 132E Baker Hall brian@stat.cmu.edu 1 Outline Bayesian Statistics and MCMC Distribution of Skill Mastery in a Population
36-463/663Multilevel and Hierarchical Models From Bayes to MCMC to MLMs Brian Junker 132E Baker Hall brian@stat.cmu.edu 1 Outline Bayesian Statistics and MCMC Distribution of Skill Mastery in a Population
Part 6: Multivariate Normal and Linear Models
 Part 6: Multivariate Normal and Linear Models 1 Multiple measurements Up until now all of our statistical models have been univariate models models for a single measurement on each member of a sample of
Part 6: Multivariate Normal and Linear Models 1 Multiple measurements Up until now all of our statistical models have been univariate models models for a single measurement on each member of a sample of
Prior. Credibility Possibilities. Posterior. B is impossible. Credibility
 Figures and Tables from Kruschke, J. K. (). Doing Bayesian Data Analysis: A Tutorial with R, JAGS, and Stan. nd Edition. Academic Press / Elsevier. This document is intended as a presentation aid for instructors
Figures and Tables from Kruschke, J. K. (). Doing Bayesian Data Analysis: A Tutorial with R, JAGS, and Stan. nd Edition. Academic Press / Elsevier. This document is intended as a presentation aid for instructors
PASS Sample Size Software. Poisson Regression
 Chapter 870 Introduction Poisson regression is used when the dependent variable is a count. Following the results of Signorini (99), this procedure calculates power and sample size for testing the hypothesis
Chapter 870 Introduction Poisson regression is used when the dependent variable is a count. Following the results of Signorini (99), this procedure calculates power and sample size for testing the hypothesis
BUGS Bayesian inference Using Gibbs Sampling
 BUGS Bayesian inference Using Gibbs Sampling Glen DePalma Department of Statistics May 30, 2013 www.stat.purdue.edu/~gdepalma 1 / 20 Bayesian Philosophy I [Pearl] turned Bayesian in 1971, as soon as I
BUGS Bayesian inference Using Gibbs Sampling Glen DePalma Department of Statistics May 30, 2013 www.stat.purdue.edu/~gdepalma 1 / 20 Bayesian Philosophy I [Pearl] turned Bayesian in 1971, as soon as I
 Weakness of Beta priors (or conjugate priors in general) They can only represent a limited range of prior beliefs. For example... There are no bimodal beta distributions (except when the modes are at 0
Weakness of Beta priors (or conjugate priors in general) They can only represent a limited range of prior beliefs. For example... There are no bimodal beta distributions (except when the modes are at 0
R Demonstration ANCOVA
 R Demonstration ANCOVA Objective: The purpose of this week s session is to demonstrate how to perform an analysis of covariance (ANCOVA) in R, and how to plot the regression lines for each level of the
R Demonstration ANCOVA Objective: The purpose of this week s session is to demonstrate how to perform an analysis of covariance (ANCOVA) in R, and how to plot the regression lines for each level of the
Generalized Linear Models for Non-Normal Data
 Generalized Linear Models for Non-Normal Data Today s Class: 3 parts of a generalized model Models for binary outcomes Complications for generalized multivariate or multilevel models SPLH 861: Lecture
Generalized Linear Models for Non-Normal Data Today s Class: 3 parts of a generalized model Models for binary outcomes Complications for generalized multivariate or multilevel models SPLH 861: Lecture
MAS3301 Bayesian Statistics Problems 5 and Solutions
 MAS3301 Bayesian Statistics Problems 5 and Solutions Semester 008-9 Problems 5 1. (Some of this question is also in Problems 4). I recorded the attendance of students at tutorials for a module. Suppose
MAS3301 Bayesian Statistics Problems 5 and Solutions Semester 008-9 Problems 5 1. (Some of this question is also in Problems 4). I recorded the attendance of students at tutorials for a module. Suppose
Lecture 19. Spatial GLM + Point Reference Spatial Data. Colin Rundel 04/03/2017
 Lecture 19 Spatial GLM + Point Reference Spatial Data Colin Rundel 04/03/2017 1 Spatial GLM Models 2 Scottish Lip Cancer Data Observed Expected 60 N 59 N 58 N 57 N 56 N value 80 60 40 20 0 55 N 8 W 6 W
Lecture 19 Spatial GLM + Point Reference Spatial Data Colin Rundel 04/03/2017 1 Spatial GLM Models 2 Scottish Lip Cancer Data Observed Expected 60 N 59 N 58 N 57 N 56 N value 80 60 40 20 0 55 N 8 W 6 W
Why Bayesian approaches? The average height of a rare plant
 Why Bayesian approaches? The average height of a rare plant Estimation and comparison of averages is an important step in many ecological analyses and demographic models. In this demonstration you will
Why Bayesian approaches? The average height of a rare plant Estimation and comparison of averages is an important step in many ecological analyses and demographic models. In this demonstration you will
Generalized linear models
 Generalized linear models Outline for today What is a generalized linear model Linear predictors and link functions Example: estimate a proportion Analysis of deviance Example: fit dose- response data
Generalized linear models Outline for today What is a generalized linear model Linear predictors and link functions Example: estimate a proportion Analysis of deviance Example: fit dose- response data
Metropolis-Hastings Algorithm
 Strength of the Gibbs sampler Metropolis-Hastings Algorithm Easy algorithm to think about. Exploits the factorization properties of the joint probability distribution. No difficult choices to be made to
Strength of the Gibbs sampler Metropolis-Hastings Algorithm Easy algorithm to think about. Exploits the factorization properties of the joint probability distribution. No difficult choices to be made to
Stat 587: Key points and formulae Week 15
 Odds ratios to compare two proportions: Difference, p 1 p 2, has issues when applied to many populations Vit. C: P[cold Placebo] = 0.82, P[cold Vit. C] = 0.74, Estimated diff. is 8% What if a year or place
Odds ratios to compare two proportions: Difference, p 1 p 2, has issues when applied to many populations Vit. C: P[cold Placebo] = 0.82, P[cold Vit. C] = 0.74, Estimated diff. is 8% What if a year or place
Multivariate Bayesian Linear Regression MLAI Lecture 11
 Multivariate Bayesian Linear Regression MLAI Lecture 11 Neil D. Lawrence Department of Computer Science Sheffield University 21st October 2012 Outline Univariate Bayesian Linear Regression Multivariate
Multivariate Bayesian Linear Regression MLAI Lecture 11 Neil D. Lawrence Department of Computer Science Sheffield University 21st October 2012 Outline Univariate Bayesian Linear Regression Multivariate
 Weakness of Beta priors (or conjugate priors in general) They can only represent a limited range of prior beliefs. For example... There are no bimodal beta distributions (except when the modes are at 0
Weakness of Beta priors (or conjugate priors in general) They can only represent a limited range of prior beliefs. For example... There are no bimodal beta distributions (except when the modes are at 0
MAS3301 Bayesian Statistics
 MAS3301 Bayesian Statistics M. Farrow School of Mathematics and Statistics Newcastle University Semester 2, 2008-9 1 11 Conjugate Priors IV: The Dirichlet distribution and multinomial observations 11.1
MAS3301 Bayesian Statistics M. Farrow School of Mathematics and Statistics Newcastle University Semester 2, 2008-9 1 11 Conjugate Priors IV: The Dirichlet distribution and multinomial observations 11.1
Theoretical and Computational Approaches to Dempster-Shafer (DS) Inference
 Theoretical and Computational Approaches to Dempster-Shafer (DS) Inference Chuanhai Liu Department of Statistics Purdue University Joint work with A. P. Dempster Dec. 11, 2006 What is DS Analysis (DSA)?
Theoretical and Computational Approaches to Dempster-Shafer (DS) Inference Chuanhai Liu Department of Statistics Purdue University Joint work with A. P. Dempster Dec. 11, 2006 What is DS Analysis (DSA)?
Addition to PGLR Chap 6
 Arizona State University From the SelectedWorks of Joseph M Hilbe August 27, 216 Addition to PGLR Chap 6 Joseph M Hilbe, Arizona State University Available at: https://works.bepress.com/joseph_hilbe/69/
Arizona State University From the SelectedWorks of Joseph M Hilbe August 27, 216 Addition to PGLR Chap 6 Joseph M Hilbe, Arizona State University Available at: https://works.bepress.com/joseph_hilbe/69/
Generalized Linear Models
 York SPIDA John Fox Notes Generalized Linear Models Copyright 2010 by John Fox Generalized Linear Models 1 1. Topics I The structure of generalized linear models I Poisson and other generalized linear
York SPIDA John Fox Notes Generalized Linear Models Copyright 2010 by John Fox Generalized Linear Models 1 1. Topics I The structure of generalized linear models I Poisson and other generalized linear
LASSO-Type Penalization in the Framework of Generalized Additive Models for Location, Scale and Shape
 LASSO-Type Penalization in the Framework of Generalized Additive Models for Location, Scale and Shape Nikolaus Umlauf https://eeecon.uibk.ac.at/~umlauf/ Overview Joint work with Andreas Groll, Julien Hambuckers
LASSO-Type Penalization in the Framework of Generalized Additive Models for Location, Scale and Shape Nikolaus Umlauf https://eeecon.uibk.ac.at/~umlauf/ Overview Joint work with Andreas Groll, Julien Hambuckers
Example using R: Heart Valves Study
 Example using R: Heart Valves Study Goal: Show that the thrombogenicity rate (TR) is less than two times the objective performance criterion R and WinBUGS Examples p. 1/27 Example using R: Heart Valves
Example using R: Heart Valves Study Goal: Show that the thrombogenicity rate (TR) is less than two times the objective performance criterion R and WinBUGS Examples p. 1/27 Example using R: Heart Valves
Logistic Regression. Interpretation of linear regression. Other types of outcomes. 0-1 response variable: Wound infection. Usual linear regression
 Logistic Regression Usual linear regression (repetition) y i = b 0 + b 1 x 1i + b 2 x 2i + e i, e i N(0,σ 2 ) or: y i N(b 0 + b 1 x 1i + b 2 x 2i,σ 2 ) Example (DGA, p. 336): E(PEmax) = 47.355 + 1.024
Logistic Regression Usual linear regression (repetition) y i = b 0 + b 1 x 1i + b 2 x 2i + e i, e i N(0,σ 2 ) or: y i N(b 0 + b 1 x 1i + b 2 x 2i,σ 2 ) Example (DGA, p. 336): E(PEmax) = 47.355 + 1.024
Generalized Linear Models 1
 Generalized Linear Models 1 STA 2101/442: Fall 2012 1 See last slide for copyright information. 1 / 24 Suggested Reading: Davison s Statistical models Exponential families of distributions Sec. 5.2 Chapter
Generalized Linear Models 1 STA 2101/442: Fall 2012 1 See last slide for copyright information. 1 / 24 Suggested Reading: Davison s Statistical models Exponential families of distributions Sec. 5.2 Chapter
BMI 541/699 Lecture 22
 BMI 541/699 Lecture 22 Where we are: 1. Introduction and Experimental Design 2. Exploratory Data Analysis 3. Probability 4. T-based methods for continous variables 5. Power and sample size for t-based
BMI 541/699 Lecture 22 Where we are: 1. Introduction and Experimental Design 2. Exploratory Data Analysis 3. Probability 4. T-based methods for continous variables 5. Power and sample size for t-based
High-Throughput Sequencing Course
 High-Throughput Sequencing Course DESeq Model for RNA-Seq Biostatistics and Bioinformatics Summer 2017 Outline Review: Standard linear regression model (e.g., to model gene expression as function of an
High-Throughput Sequencing Course DESeq Model for RNA-Seq Biostatistics and Bioinformatics Summer 2017 Outline Review: Standard linear regression model (e.g., to model gene expression as function of an
HW3 Solutions : Applied Bayesian and Computational Statistics
 HW3 Solutions 36-724: Applied Bayesian and Computational Statistics March 2, 2006 Problem 1 a Fatal Accidents Poisson(θ I will set a prior for θ to be Gamma, as it is the conjugate prior. I will allow
HW3 Solutions 36-724: Applied Bayesian and Computational Statistics March 2, 2006 Problem 1 a Fatal Accidents Poisson(θ I will set a prior for θ to be Gamma, as it is the conjugate prior. I will allow
Part 8: GLMs and Hierarchical LMs and GLMs
 Part 8: GLMs and Hierarchical LMs and GLMs 1 Example: Song sparrow reproductive success Arcese et al., (1992) provide data on a sample from a population of 52 female song sparrows studied over the course
Part 8: GLMs and Hierarchical LMs and GLMs 1 Example: Song sparrow reproductive success Arcese et al., (1992) provide data on a sample from a population of 52 female song sparrows studied over the course
The Jeffreys Prior. Yingbo Li MATH Clemson University. Yingbo Li (Clemson) The Jeffreys Prior MATH / 13
 The Jeffreys Prior Yingbo Li Clemson University MATH 9810 Yingbo Li (Clemson) The Jeffreys Prior MATH 9810 1 / 13 Sir Harold Jeffreys English mathematician, statistician, geophysicist, and astronomer His
The Jeffreys Prior Yingbo Li Clemson University MATH 9810 Yingbo Li (Clemson) The Jeffreys Prior MATH 9810 1 / 13 Sir Harold Jeffreys English mathematician, statistician, geophysicist, and astronomer His
Regression models. Generalized linear models in R. Normal regression models are not always appropriate. Generalized linear models. Examples.
 Regression models Generalized linear models in R Dr Peter K Dunn http://www.usq.edu.au Department of Mathematics and Computing University of Southern Queensland ASC, July 00 The usual linear regression
Regression models Generalized linear models in R Dr Peter K Dunn http://www.usq.edu.au Department of Mathematics and Computing University of Southern Queensland ASC, July 00 The usual linear regression
Hypothesis Testing: Chi-Square Test 1
 Hypothesis Testing: Chi-Square Test 1 November 9, 2017 1 HMS, 2017, v1.0 Chapter References Diez: Chapter 6.3 Navidi, Chapter 6.10 Chapter References 2 Chi-square Distributions Let X 1, X 2,... X n be
Hypothesis Testing: Chi-Square Test 1 November 9, 2017 1 HMS, 2017, v1.0 Chapter References Diez: Chapter 6.3 Navidi, Chapter 6.10 Chapter References 2 Chi-square Distributions Let X 1, X 2,... X n be
NATIONAL UNIVERSITY OF SINGAPORE EXAMINATION. ST3241 Categorical Data Analysis. (Semester II: ) April/May, 2011 Time Allowed : 2 Hours
 NATIONAL UNIVERSITY OF SINGAPORE EXAMINATION Categorical Data Analysis (Semester II: 2010 2011) April/May, 2011 Time Allowed : 2 Hours Matriculation No: Seat No: Grade Table Question 1 2 3 4 5 6 Full marks
NATIONAL UNIVERSITY OF SINGAPORE EXAMINATION Categorical Data Analysis (Semester II: 2010 2011) April/May, 2011 Time Allowed : 2 Hours Matriculation No: Seat No: Grade Table Question 1 2 3 4 5 6 Full marks
The evdbayes Package
 The evdbayes Package April 19, 2006 Version 1.0-5 Date 2006-18-04 Title Bayesian Analysis in Extreme Theory Author Alec Stephenson and Mathieu Ribatet. Maintainer Mathieu Ribatet
The evdbayes Package April 19, 2006 Version 1.0-5 Date 2006-18-04 Title Bayesian Analysis in Extreme Theory Author Alec Stephenson and Mathieu Ribatet. Maintainer Mathieu Ribatet
Stat 5421 Lecture Notes Proper Conjugate Priors for Exponential Families Charles J. Geyer March 28, 2016
 Stat 5421 Lecture Notes Proper Conjugate Priors for Exponential Families Charles J. Geyer March 28, 2016 1 Theory This section explains the theory of conjugate priors for exponential families of distributions,
Stat 5421 Lecture Notes Proper Conjugate Priors for Exponential Families Charles J. Geyer March 28, 2016 1 Theory This section explains the theory of conjugate priors for exponential families of distributions,
Linear Regression Models P8111
 Linear Regression Models P8111 Lecture 25 Jeff Goldsmith April 26, 2016 1 of 37 Today s Lecture Logistic regression / GLMs Model framework Interpretation Estimation 2 of 37 Linear regression Course started
Linear Regression Models P8111 Lecture 25 Jeff Goldsmith April 26, 2016 1 of 37 Today s Lecture Logistic regression / GLMs Model framework Interpretation Estimation 2 of 37 Linear regression Course started
Contents. 1 Introduction: what is overdispersion? 2 Recognising (and testing for) overdispersion. 1 Introduction: what is overdispersion?
 Overdispersion, and how to deal with it in R and JAGS (requires R-packages AER, coda, lme4, R2jags, DHARMa/devtools) Carsten F. Dormann 07 December, 2016 Contents 1 Introduction: what is overdispersion?
Overdispersion, and how to deal with it in R and JAGS (requires R-packages AER, coda, lme4, R2jags, DHARMa/devtools) Carsten F. Dormann 07 December, 2016 Contents 1 Introduction: what is overdispersion?
Ph.D. Qualifying Exam Friday Saturday, January 6 7, 2017
 Ph.D. Qualifying Exam Friday Saturday, January 6 7, 2017 Put your solution to each problem on a separate sheet of paper. Problem 1. (5106) Let X 1, X 2,, X n be a sequence of i.i.d. observations from a
Ph.D. Qualifying Exam Friday Saturday, January 6 7, 2017 Put your solution to each problem on a separate sheet of paper. Problem 1. (5106) Let X 1, X 2,, X n be a sequence of i.i.d. observations from a
Hierarchical Generalized Linear Models. ERSH 8990 REMS Seminar on HLM Last Lecture!
 Hierarchical Generalized Linear Models ERSH 8990 REMS Seminar on HLM Last Lecture! Hierarchical Generalized Linear Models Introduction to generalized models Models for binary outcomes Interpreting parameter
Hierarchical Generalized Linear Models ERSH 8990 REMS Seminar on HLM Last Lecture! Hierarchical Generalized Linear Models Introduction to generalized models Models for binary outcomes Interpreting parameter
Stat 5101 Lecture Notes
 Stat 5101 Lecture Notes Charles J. Geyer Copyright 1998, 1999, 2000, 2001 by Charles J. Geyer May 7, 2001 ii Stat 5101 (Geyer) Course Notes Contents 1 Random Variables and Change of Variables 1 1.1 Random
Stat 5101 Lecture Notes Charles J. Geyer Copyright 1998, 1999, 2000, 2001 by Charles J. Geyer May 7, 2001 ii Stat 5101 (Geyer) Course Notes Contents 1 Random Variables and Change of Variables 1 1.1 Random
12 Generalized linear models
 12 Generalized linear models In this chapter, we combine regression models with other parametric probability models like the binomial and Poisson distributions. Binary responses In many situations, we
12 Generalized linear models In this chapter, we combine regression models with other parametric probability models like the binomial and Poisson distributions. Binary responses In many situations, we
Here θ = (α, β) is the unknown parameter. The likelihood function is
 Stat 591 Notes Logistic regression and Metropolis Hastings example Ryan Martin (rgmartin@uic.edu) November 2, 2013 Introduction (This part taken from Example 1.13 in Robert and Casella, 2004.) In 1986,
Stat 591 Notes Logistic regression and Metropolis Hastings example Ryan Martin (rgmartin@uic.edu) November 2, 2013 Introduction (This part taken from Example 1.13 in Robert and Casella, 2004.) In 1986,
Varieties of Count Data
 CHAPTER 1 Varieties of Count Data SOME POINTS OF DISCUSSION What are counts? What are count data? What is a linear statistical model? What is the relationship between a probability distribution function
CHAPTER 1 Varieties of Count Data SOME POINTS OF DISCUSSION What are counts? What are count data? What is a linear statistical model? What is the relationship between a probability distribution function
Generalized Linear Models and Exponential Families
 Generalized Linear Models and Exponential Families David M. Blei COS424 Princeton University April 12, 2012 Generalized Linear Models x n y n β Linear regression and logistic regression are both linear
Generalized Linear Models and Exponential Families David M. Blei COS424 Princeton University April 12, 2012 Generalized Linear Models x n y n β Linear regression and logistic regression are both linear
GOV 2001/ 1002/ E-2001 Section 3 Theories of Inference
 GOV 2001/ 1002/ E-2001 Section 3 Theories of Inference Solé Prillaman Harvard University February 11, 2015 1 / 48 LOGISTICS Reading Assignment- Unifying Political Methodology chs 2 and 4. Problem Set 3-
GOV 2001/ 1002/ E-2001 Section 3 Theories of Inference Solé Prillaman Harvard University February 11, 2015 1 / 48 LOGISTICS Reading Assignment- Unifying Political Methodology chs 2 and 4. Problem Set 3-
Package LBLGXE. R topics documented: July 20, Type Package
 Type Package Package LBLGXE July 20, 2015 Title Bayesian Lasso for detecting Rare (or Common) Haplotype Association and their interactions with Environmental Covariates Version 1.2 Date 2015-07-09 Author
Type Package Package LBLGXE July 20, 2015 Title Bayesian Lasso for detecting Rare (or Common) Haplotype Association and their interactions with Environmental Covariates Version 1.2 Date 2015-07-09 Author
ST417 Introduction to Bayesian Modelling. Conjugate Modelling (Poisson-Gamma)
 ST417 Introduction to Bayesian Modelling Conjugate Modelling (Poisson-Gamma) Slides based on the source provided courtesy of Prof. D.Draper, UCSC ST417 Intro to Bayesian Modelling 1 Integer-Valued Outcomes
ST417 Introduction to Bayesian Modelling Conjugate Modelling (Poisson-Gamma) Slides based on the source provided courtesy of Prof. D.Draper, UCSC ST417 Intro to Bayesian Modelling 1 Integer-Valued Outcomes
Lecture 14: Introduction to Poisson Regression
 Lecture 14: Introduction to Poisson Regression Ani Manichaikul amanicha@jhsph.edu 8 May 2007 1 / 52 Overview Modelling counts Contingency tables Poisson regression models 2 / 52 Modelling counts I Why
Lecture 14: Introduction to Poisson Regression Ani Manichaikul amanicha@jhsph.edu 8 May 2007 1 / 52 Overview Modelling counts Contingency tables Poisson regression models 2 / 52 Modelling counts I Why
Modelling counts. Lecture 14: Introduction to Poisson Regression. Overview
 Modelling counts I Lecture 14: Introduction to Poisson Regression Ani Manichaikul amanicha@jhsph.edu Why count data? Number of traffic accidents per day Mortality counts in a given neighborhood, per week
Modelling counts I Lecture 14: Introduction to Poisson Regression Ani Manichaikul amanicha@jhsph.edu Why count data? Number of traffic accidents per day Mortality counts in a given neighborhood, per week
Subject CS1 Actuarial Statistics 1 Core Principles
 Institute of Actuaries of India Subject CS1 Actuarial Statistics 1 Core Principles For 2019 Examinations Aim The aim of the Actuarial Statistics 1 subject is to provide a grounding in mathematical and
Institute of Actuaries of India Subject CS1 Actuarial Statistics 1 Core Principles For 2019 Examinations Aim The aim of the Actuarial Statistics 1 subject is to provide a grounding in mathematical and
22s:152 Applied Linear Regression. Example: Study on lead levels in children. Ch. 14 (sec. 1) and Ch. 15 (sec. 1 & 4): Logistic Regression
 22s:52 Applied Linear Regression Ch. 4 (sec. and Ch. 5 (sec. & 4: Logistic Regression Logistic Regression When the response variable is a binary variable, such as 0 or live or die fail or succeed then
22s:52 Applied Linear Regression Ch. 4 (sec. and Ch. 5 (sec. & 4: Logistic Regression Logistic Regression When the response variable is a binary variable, such as 0 or live or die fail or succeed then
Bayesian Model Diagnostics and Checking
 Earvin Balderama Quantitative Ecology Lab Department of Forestry and Environmental Resources North Carolina State University April 12, 2013 1 / 34 Introduction MCMCMC 2 / 34 Introduction MCMCMC Steps in
Earvin Balderama Quantitative Ecology Lab Department of Forestry and Environmental Resources North Carolina State University April 12, 2013 1 / 34 Introduction MCMCMC 2 / 34 Introduction MCMCMC Steps in
Statistical techniques for data analysis in Cosmology
 Statistical techniques for data analysis in Cosmology arxiv:0712.3028; arxiv:0911.3105 Numerical recipes (the bible ) Licia Verde ICREA & ICC UB-IEEC http://icc.ub.edu/~liciaverde outline Lecture 1: Introduction
Statistical techniques for data analysis in Cosmology arxiv:0712.3028; arxiv:0911.3105 Numerical recipes (the bible ) Licia Verde ICREA & ICC UB-IEEC http://icc.ub.edu/~liciaverde outline Lecture 1: Introduction
SPRING 2007 EXAM C SOLUTIONS
 SPRING 007 EXAM C SOLUTIONS Question #1 The data are already shifted (have had the policy limit and the deductible of 50 applied). The two 350 payments are censored. Thus the likelihood function is L =
SPRING 007 EXAM C SOLUTIONS Question #1 The data are already shifted (have had the policy limit and the deductible of 50 applied). The two 350 payments are censored. Thus the likelihood function is L =
The lmm Package. May 9, Description Some improved procedures for linear mixed models
 The lmm Package May 9, 2005 Version 0.3-4 Date 2005-5-9 Title Linear mixed models Author Original by Joseph L. Schafer . Maintainer Jing hua Zhao Description Some improved
The lmm Package May 9, 2005 Version 0.3-4 Date 2005-5-9 Title Linear mixed models Author Original by Joseph L. Schafer . Maintainer Jing hua Zhao Description Some improved
Today. Calculus. Linear Regression. Lagrange Multipliers
 Today Calculus Lagrange Multipliers Linear Regression 1 Optimization with constraints What if I want to constrain the parameters of the model. The mean is less than 10 Find the best likelihood, subject
Today Calculus Lagrange Multipliers Linear Regression 1 Optimization with constraints What if I want to constrain the parameters of the model. The mean is less than 10 Find the best likelihood, subject
A New Bayesian Variable Selection Method: The Bayesian Lasso with Pseudo Variables
 A New Bayesian Variable Selection Method: The Bayesian Lasso with Pseudo Variables Qi Tang (Joint work with Kam-Wah Tsui and Sijian Wang) Department of Statistics University of Wisconsin-Madison Feb. 8,
A New Bayesian Variable Selection Method: The Bayesian Lasso with Pseudo Variables Qi Tang (Joint work with Kam-Wah Tsui and Sijian Wang) Department of Statistics University of Wisconsin-Madison Feb. 8,
Stat 5421 Lecture Notes Simple Chi-Square Tests for Contingency Tables Charles J. Geyer March 12, 2016
 Stat 5421 Lecture Notes Simple Chi-Square Tests for Contingency Tables Charles J. Geyer March 12, 2016 1 One-Way Contingency Table The data set read in by the R function read.table below simulates 6000
Stat 5421 Lecture Notes Simple Chi-Square Tests for Contingency Tables Charles J. Geyer March 12, 2016 1 One-Way Contingency Table The data set read in by the R function read.table below simulates 6000
1. Hypothesis testing through analysis of deviance. 3. Model & variable selection - stepwise aproaches
 Sta 216, Lecture 4 Last Time: Logistic regression example, existence/uniqueness of MLEs Today s Class: 1. Hypothesis testing through analysis of deviance 2. Standard errors & confidence intervals 3. Model
Sta 216, Lecture 4 Last Time: Logistic regression example, existence/uniqueness of MLEs Today s Class: 1. Hypothesis testing through analysis of deviance 2. Standard errors & confidence intervals 3. Model
Correspondence Analysis
 Correspondence Analysis Q: when independence of a 2-way contingency table is rejected, how to know where the dependence is coming from? The interaction terms in a GLM contain dependence information; however,
Correspondence Analysis Q: when independence of a 2-way contingency table is rejected, how to know where the dependence is coming from? The interaction terms in a GLM contain dependence information; however,
10. Exchangeability and hierarchical models Objective. Recommended reading
 10. Exchangeability and hierarchical models Objective Introduce exchangeability and its relation to Bayesian hierarchical models. Show how to fit such models using fully and empirical Bayesian methods.
10. Exchangeability and hierarchical models Objective Introduce exchangeability and its relation to Bayesian hierarchical models. Show how to fit such models using fully and empirical Bayesian methods.
Binary Logistic Regression
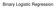 The coefficients of the multiple regression model are estimated using sample data with k independent variables Estimated (or predicted) value of Y Estimated intercept Estimated slope coefficients Ŷ = b
The coefficients of the multiple regression model are estimated using sample data with k independent variables Estimated (or predicted) value of Y Estimated intercept Estimated slope coefficients Ŷ = b
Heriot-Watt University
 Heriot-Watt University Heriot-Watt University Research Gateway Prediction of settlement delay in critical illness insurance claims by using the generalized beta of the second kind distribution Dodd, Erengul;
Heriot-Watt University Heriot-Watt University Research Gateway Prediction of settlement delay in critical illness insurance claims by using the generalized beta of the second kind distribution Dodd, Erengul;
Lecture 16 : Bayesian analysis of contingency tables. Bayesian linear regression. Jonathan Marchini (University of Oxford) BS2a MT / 15
 Lecture 16 : Bayesian analysis of contingency tables. Bayesian linear regression. Jonathan Marchini (University of Oxford) BS2a MT 2013 1 / 15 Contingency table analysis North Carolina State University
Lecture 16 : Bayesian analysis of contingency tables. Bayesian linear regression. Jonathan Marchini (University of Oxford) BS2a MT 2013 1 / 15 Contingency table analysis North Carolina State University
Modelling geoadditive survival data
 Modelling geoadditive survival data Thomas Kneib & Ludwig Fahrmeir Department of Statistics, Ludwig-Maximilians-University Munich 1. Leukemia survival data 2. Structured hazard regression 3. Mixed model
Modelling geoadditive survival data Thomas Kneib & Ludwig Fahrmeir Department of Statistics, Ludwig-Maximilians-University Munich 1. Leukemia survival data 2. Structured hazard regression 3. Mixed model
Generalized Models: Part 1
 Generalized Models: Part 1 Topics: Introduction to generalized models Introduction to maximum likelihood estimation Models for binary outcomes Models for proportion outcomes Models for categorical outcomes
Generalized Models: Part 1 Topics: Introduction to generalized models Introduction to maximum likelihood estimation Models for binary outcomes Models for proportion outcomes Models for categorical outcomes
STA 216, GLM, Lecture 16. October 29, 2007
 STA 216, GLM, Lecture 16 October 29, 2007 Efficient Posterior Computation in Factor Models Underlying Normal Models Generalized Latent Trait Models Formulation Genetic Epidemiology Illustration Structural
STA 216, GLM, Lecture 16 October 29, 2007 Efficient Posterior Computation in Factor Models Underlying Normal Models Generalized Latent Trait Models Formulation Genetic Epidemiology Illustration Structural
Glossary. The ISI glossary of statistical terms provides definitions in a number of different languages:
 Glossary The ISI glossary of statistical terms provides definitions in a number of different languages: http://isi.cbs.nl/glossary/index.htm Adjusted r 2 Adjusted R squared measures the proportion of the
Glossary The ISI glossary of statistical terms provides definitions in a number of different languages: http://isi.cbs.nl/glossary/index.htm Adjusted r 2 Adjusted R squared measures the proportion of the
Generalized logit models for nominal multinomial responses. Local odds ratios
 Generalized logit models for nominal multinomial responses Categorical Data Analysis, Summer 2015 1/17 Local odds ratios Y 1 2 3 4 1 π 11 π 12 π 13 π 14 π 1+ X 2 π 21 π 22 π 23 π 24 π 2+ 3 π 31 π 32 π
Generalized logit models for nominal multinomial responses Categorical Data Analysis, Summer 2015 1/17 Local odds ratios Y 1 2 3 4 1 π 11 π 12 π 13 π 14 π 1+ X 2 π 21 π 22 π 23 π 24 π 2+ 3 π 31 π 32 π
Bayes methods for categorical data. April 25, 2017
 Bayes methods for categorical data April 25, 2017 Motivation for joint probability models Increasing interest in high-dimensional data in broad applications Focus may be on prediction, variable selection,
Bayes methods for categorical data April 25, 2017 Motivation for joint probability models Increasing interest in high-dimensional data in broad applications Focus may be on prediction, variable selection,
Bayesian Networks in Educational Assessment
 Bayesian Networks in Educational Assessment Estimating Parameters with MCMC Bayesian Inference: Expanding Our Context Roy Levy Arizona State University Roy.Levy@asu.edu 2017 Roy Levy MCMC 1 MCMC 2 Posterior
Bayesian Networks in Educational Assessment Estimating Parameters with MCMC Bayesian Inference: Expanding Our Context Roy Levy Arizona State University Roy.Levy@asu.edu 2017 Roy Levy MCMC 1 MCMC 2 Posterior
10702/36702 Statistical Machine Learning, Spring 2008: Homework 3 Solutions
 10702/36702 Statistical Machine Learning, Spring 2008: Homework 3 Solutions March 24, 2008 1 [25 points], (Jingrui) (a) Generate data as follows. n = 100 p = 1000 X = matrix(rnorm(n p), n, p) beta = c(rep(10,
10702/36702 Statistical Machine Learning, Spring 2008: Homework 3 Solutions March 24, 2008 1 [25 points], (Jingrui) (a) Generate data as follows. n = 100 p = 1000 X = matrix(rnorm(n p), n, p) beta = c(rep(10,
Poisson Regression. Gelman & Hill Chapter 6. February 6, 2017
 Poisson Regression Gelman & Hill Chapter 6 February 6, 2017 Military Coups Background: Sub-Sahara Africa has experienced a high proportion of regime changes due to military takeover of governments for
Poisson Regression Gelman & Hill Chapter 6 February 6, 2017 Military Coups Background: Sub-Sahara Africa has experienced a high proportion of regime changes due to military takeover of governments for
Chapter 22: Log-linear regression for Poisson counts
 Chapter 22: Log-linear regression for Poisson counts Exposure to ionizing radiation is recognized as a cancer risk. In the United States, EPA sets guidelines specifying upper limits on the amount of exposure
Chapter 22: Log-linear regression for Poisson counts Exposure to ionizing radiation is recognized as a cancer risk. In the United States, EPA sets guidelines specifying upper limits on the amount of exposure
Introduction to the Generalized Linear Model: Logistic regression and Poisson regression
 Introduction to the Generalized Linear Model: Logistic regression and Poisson regression Statistical modelling: Theory and practice Gilles Guillot gigu@dtu.dk November 4, 2013 Gilles Guillot (gigu@dtu.dk)
Introduction to the Generalized Linear Model: Logistic regression and Poisson regression Statistical modelling: Theory and practice Gilles Guillot gigu@dtu.dk November 4, 2013 Gilles Guillot (gigu@dtu.dk)
Final Exam. Name: Solution:
 Final Exam. Name: Instructions. Answer all questions on the exam. Open books, open notes, but no electronic devices. The first 13 problems are worth 5 points each. The rest are worth 1 point each. HW1.
Final Exam. Name: Instructions. Answer all questions on the exam. Open books, open notes, but no electronic devices. The first 13 problems are worth 5 points each. The rest are worth 1 point each. HW1.
Tento projekt je spolufinancován Evropským sociálním fondem a Státním rozpočtem ČR InoBio CZ.1.07/2.2.00/
 Tento projekt je spolufinancován Evropským sociálním fondem a Státním rozpočtem ČR InoBio CZ.1.07/2.2.00/28.0018 Statistical Analysis in Ecology using R Linear Models/GLM Ing. Daniel Volařík, Ph.D. 13.
Tento projekt je spolufinancován Evropským sociálním fondem a Státním rozpočtem ČR InoBio CZ.1.07/2.2.00/28.0018 Statistical Analysis in Ecology using R Linear Models/GLM Ing. Daniel Volařík, Ph.D. 13.
Class Notes: Week 8. Probit versus Logit Link Functions and Count Data
 Ronald Heck Class Notes: Week 8 1 Class Notes: Week 8 Probit versus Logit Link Functions and Count Data This week we ll take up a couple of issues. The first is working with a probit link function. While
Ronald Heck Class Notes: Week 8 1 Class Notes: Week 8 Probit versus Logit Link Functions and Count Data This week we ll take up a couple of issues. The first is working with a probit link function. While
The Exciting Guide To Probability Distributions Part 2. Jamie Frost v1.1
 The Exciting Guide To Probability Distributions Part 2 Jamie Frost v. Contents Part 2 A revisit of the multinomial distribution The Dirichlet Distribution The Beta Distribution Conjugate Priors The Gamma
The Exciting Guide To Probability Distributions Part 2 Jamie Frost v. Contents Part 2 A revisit of the multinomial distribution The Dirichlet Distribution The Beta Distribution Conjugate Priors The Gamma
Statistical Models with Uncertain Error Parameters (G. Cowan, arxiv: )
 Statistical Models with Uncertain Error Parameters (G. Cowan, arxiv:1809.05778) Workshop on Advanced Statistics for Physics Discovery aspd.stat.unipd.it Department of Statistical Sciences, University of
Statistical Models with Uncertain Error Parameters (G. Cowan, arxiv:1809.05778) Workshop on Advanced Statistics for Physics Discovery aspd.stat.unipd.it Department of Statistical Sciences, University of
