Quantifying the relevance of intraspecific trait variability for functional diversity
|
|
|
- Lindsey Bates
- 5 years ago
- Views:
Transcription
1 Tutorial Quantifying the relevance of intraspecific trait variability for functional diversity Francesco de Bello, Sandra Lavorel, Cécile H. Albert, Wilfried Thuiller, Karl Grigulis, Jiři Dolezal, Štěpán Janeček and Jan Lepš
2 Intraspecific trait variability is a crucial, often neglected, component of functional diversity (i.e. the extent of trait dissimilarity in a given ecological community). e.g. traits = plant height, n of leaves, n of flowers... How much trait variability is due to intraspecific variation within and across communities?
3 Two new methods proposed 1 Quantifying the extent of within- vs. between-species functional diversity (FD) within different communities 2 Decomposing the effects of species turnover and intraspecific trait variability in FD across communities
4 Present TUTORIAL: Use of the R functions designed for the first method 1 Quantifying the extent of within- vs. between-species functional diversity (FD) within different communities 2 Decomposing the effects of species turnover and intraspecific trait variability in FD across communities Method overview Data preparation Running the R functions Reading the results
5 Within each community (plot) the functional trait diversity can be partitioned into effects due to: Between species FD: extent of trait dissimilarity in a community because of differentiation between coexisting species Vs. Within species FD: extent of trait dissimilarity in a community because of intraspecific trait variability Vs. Method overview
6 APPROACH: decomposition of total community trait variance Total diversity = Between species div. + Within species div. ADVANTAGES Take into account different type of species abundances Similar to the decomposition with the Rao quadratic entropy Similar to PERMANOVA and other existing mathematical tools Use with single and multiple traits Method overview
7 Total diversity = Between species div. + Within species div. Calculated for each community (plot) separately!! Functional diversity Within sp. Between sp. Total Method overview Comm1 Comm2 Comm3 Example: Comm2 has lowest within species diversity, but highest between species and total diversity
8 Total diversity = Between species div. + Within species div. 3 possible ways to calculate the weight of each species (see Eqn. 3 in the paper) R scripts (in the paper Appendix) 1 all species have the same contribution (i.e. 1/nsp, nsp=number of species in a plot) 2 species contribution is given by their abundance in classic relevés (cover, biomass etc..) RaoRel.r 3 species contribution is given by the number of individuals sampled for trait measurements RaoAdo.r Method overview
9 Preparing the data for R 3 matrices needed (FOR EACH COMMUNITY/PLOT): 1 Trait values for different individuals MATRICES 2a 3 2b Individual identity (with and without species abundances) Trait dissimilarity (computed from the first matrix) Data preparation
10 Preparing the data for R 3 matrices needed (FOR EACH COMMUNITY/PLOT): The following examples are based on virtual data as used in the Appendix example The R functions are applicable to your real data when formatted as shown in the virtual examples Data preparation
11 1 TRAIT X INDIVIDUAL For each trait (e.g. plant height, leaf area etc..), a vector with the trait value for different individuals within each species Example: for plant height, 6 individuals were measured for species 1, 8 individuals for species 2, 10 for species 3 and 16 for species 4. Data preparation
12 Matrix 2a SPECIES IDENTITY This matrix denotes species identity of each individual collected, as a dummy code. Data preparation MATRIX FOR EACH COMMUNITY SAMPLED Species Sp1 Sp2 Sp3 Sp4 ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind9 for Sp ind10 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind9 for Sp ind10 for Sp ind11 for Sp ind12 for Sp ind13 for Sp ind14 for Sp ind15 for Sp ind16 for Sp
13 SPECIES IDENTITY with ABUNDANCE Sometimes species abundances in the community are available (i.e. measured independently via classic relevés or plot record). Species Sp1 Sp2 Sp3 Sp4 Matrix ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp b ind5 for Sp ind6 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp For computation purposes we ind6 for Sp ind7 for Sp allocate species abundance ind8 for Sp ind9 for Sp measured in the community/plot to ind10 for Sp ind1 for Sp ind2 for Sp all individuals of that particular ind3 for Sp ind4 for Sp species. ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind9 for Sp ind10 for Sp Example: the abundance could be estimated by ind11 for Sp ind12 for Sp species biomass (species 1= 10 g, species 2=20 g, ind14 for Sp ind13 for Sp ind15 for Sp species 3 = 30 g, species 4 = 40 g) ind16 for Sp Data preparation
14 TRAIT X INDIVIDUAL Matrix 1 EUCLIDIAN DISTANCE* Data preparation TRAIT DISSIMILARITIES between individuals *for multiple traits, the traits needs to be normalized first (see details in the paper) Matrix Species ind1 for Sp1 ind2 for Sp1 1 ind3 for Sp1 2 1 ind4 for Sp ind5 for Sp ind6 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind9 for Sp ind10 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind9 for Sp ind10 for Sp ind11 for Sp ind12 for Sp ind13 for Sp ind14 for Sp ind15 for Sp ind16 for Sp
15 Running the R functions REQUIREMENTS: - Basic knowledge of R - Having installed the package ade4 - Add the R functions to the specified working directory -(having read the paper) Running the R functions
16 R functions 1. Data set for a VIRTUAL example (as in the Appendix) > tind [1] [16] [31] > TRAIT X INDIVIDUAL Matrix object tind as for matrix 1 before 1 NOTE In the Appendix the object tind is built artificially by the commands: > tind1<-c(1:6) > tind2<-c(2:9) > tind3<-c(1:10) > tind4<-c(c(tind2*2), c(tind2*2)-2) > tind<-c(tind1, tind2, tind3, tind4) or INSTEAD IMPORT YOUR REAL MATRIX 1 Running the R functions
17 R functions > indxsp [,1] [,2] [,3] [,4] [1,] [2,] [3,] [4,] [5,] [6,] [7,] [8,] [9,] [10,] [11,] [12,] [13,] [14,] [15,] [16,] [17,] [18,] [19,] [20,] [21,] [22,] [23,] [24,] [25,] [26,] [27,] [28,] [29,] [30,] [31,] [32,] [33,] [34,] [35,] [36,] [37,] [38,] [39,] [40,] > SPECIES IDENTITY object indxsp (without relative abundance) as matrix 2a before Matrix 2a 1bis. VIRTUAL data set for the example Running the R functions
18 R functions > indxsp [,1] [,2] [,3] [,4] [1,] [2,] [3,] [4,] [5,] [6,] [7,] [8,] [9,] [10,] [11,] [12,] [13,] [14,] [15,] [16,] [17,] [18,] [19,] [20,] [21,] [22,] [23,] [24,] [25,] [26,] [27,] [28,] [29,] [30,] [31,] [32,] [33,] [34,] [35,] [36,] [37,] [38,] [39,] [40,] > NOTE 1bis. VIRTUAL data set for the example In the Appendix the object indxsp is built artificially by the commands: > indxsp<-matrix(0, 40, 4) > indxsp[1:6, 1]=1 > indxsp[7:14, 2]=1 > indxsp[15:24, 3]=1 > indxsp[25:40, 4]=1 > or INSTEAD IMPORT YOUR MATRIX 2 Running the R functions
19 R functions 2a. Calculate trait dissimilarity, open and run the RaoRel.r function 3 Trait Euclidian distances Matrix Note that to obtain the equivalence between the Rao index and Variance, the trait dissimilarity needs to be squared and divided by 2 (see paper) Running the R functions SPECIES IDENTITY object indxsp > d<-as.matrix(dist(tind)) > > source("raorel.r") > library(ade4) > Raotind<-RaoRel(sample=indxsp, dfunc=d^2/2, dphyl=null, weight=f, Jost=F, structure=null) > Matrix 2a
20 R functions 2b. Alternatively a matrix considering species abundance (as matrix 2b before) can be used with RaoRel.r SPECIES IDENTITY object indxspp (with abundance) Matrix 2b > Raotindp<-RaoRel(sample=indxspp, dfunc=d^2/2, dphyl=null, weight=t, Jost=F, structure=null) > Note the option weight needs to be specified otherwise the species abundance is not taken into account Running the R functions Species Sp1 Sp2 Sp3 Sp4 ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind9 for Sp ind10 for Sp ind1 for Sp ind2 for Sp ind3 for Sp ind4 for Sp ind5 for Sp ind6 for Sp ind7 for Sp ind8 for Sp ind9 for Sp ind10 for Sp ind11 for Sp ind12 for Sp ind13 for Sp ind14 for Sp ind15 for Sp ind16 for Sp
21 R functions 3. Run function RaoAdo.r SPECIES IDENTITY object indxsp no species abundances! Matrix 2a > source("raoado.r") > RaoPerm<-RaoAdo(sample=indxsp, dfunc=d^2/2, dphyl=null, weight=t, Jost=F, structure=null) > Note the option weight needs to be specified This way the weight of each species corresponds to the number of individuals sampled Running the R functions
22 The objects created by the functions RaoRel.r and RaoAdo.r (i.e. Raoind, Raoindp and RaoPerm; see previous pages) are LISTS where different results are stored (under xxx$fd ) Example with the Raoind object > names(raotind$fd) [1] "Mean_Alpha" "Alpha" "Gamma" "Beta_add" [5] "Beta_prop" "Pairwise_samples" > Reading the results
23 > names(raotind$fd) [1] "Mean_Alpha" "Alpha" "Gamma" "Beta_add" [5] "Beta_prop" "Pairwise_samples" > Gamma= TOTAL community diversity (within species + between species) Mean alpha = overall community WITHIN species FD Alpha = trait diversity within each species Beta_add = community BETWEEN species FD Beta_prop = proportion accounted by community BETWEEN species FD Reading the results
24 > names(raotind$fd) [1] "Mean_Alpha" "Alpha" "Gamma" "Beta_add" [5] "Beta_prop" "Pairwise_samples" > witrao<-raotind$fd$mean_alpha > witrao [1] > betrao<-raotind$fd$beta_add > betrao [1] > totrao<-raotind$fd$gamma > totrao Total diversity = Between species div. + Within species div. [1] > (betrao+witrao)==totrao [1] TRUE IT WORKS! > Raotind$FD$Beta_prop [1] > Raotind$FD$Alpha [1] > Reading the results
25 > names(raotind$fd) [1] "Mean_Alpha" "Alpha" "Gamma" "Beta_add" [5] "Beta_prop" "Pairwise_samples" > witrao<-raotind$fd$mean_alpha > witrao [1] > betrao<-raotind$fd$beta_add > betrao [1] > totrao<-raotind$fd$gamma > totrao [1] > (betrao+witrao)==totrao [1] TRUE > Raotind$FD$Beta_prop [1] > Raotind$FD$Alpha [1] > The between-species FD is lower (37%) than the within species (63%) Reading the results
26 > names(raotind$fd) [1] "Mean_Alpha" "Alpha" "Gamma" "Beta_add" [5] "Beta_prop" "Pairwise_samples" > witrao<-raotind$fd$mean_alpha > witrao [1] > betrao<-raotind$fd$beta_add > betrao [1] > totrao<-raotind$fd$gamma > totrao [1] > (betrao+witrao)==totrao [1] TRUE > Raotind$FD$Beta_prop [1] > Raotind$FD$Alpha [1] > The within species trait variance is highest for species 4 Reading the results
27 Example from the paper 1. The within species effect on community FD is NOT negligible 2. It changes across traits (height and LDMC), while considering species abundances and across different environmental conditions (e.g. dry vs. wet meadows) Reading the results
28 Tutorial Quantifying the relevance of intraspecific trait variability for functional diversity For more information, questions & suggestions:
Mesoscopic analysis of ecological networks using Hill numbers
 Mesoscopic analysis of ecological networks using Hill numbers Marc Ohlmann PhD supervisor : Wilfried Thuiller Jump In Jackson Pollock Introduction Examples and simulations Conclusion Aim : assess the diversity
Mesoscopic analysis of ecological networks using Hill numbers Marc Ohlmann PhD supervisor : Wilfried Thuiller Jump In Jackson Pollock Introduction Examples and simulations Conclusion Aim : assess the diversity
On the importance of intraspecific variability for the quantification of functional diversity
 Oikos 000: 001 011, 2011 doi: 10.1111/j.1600-0706.2011.19672.x 2011 The Authors. Oikos 2011 Nordic Society Oikos Subject Editor: Heikki Setälä. Accepted 9 May 2011 On the importance of intraspecific variability
Oikos 000: 001 011, 2011 doi: 10.1111/j.1600-0706.2011.19672.x 2011 The Authors. Oikos 2011 Nordic Society Oikos Subject Editor: Heikki Setälä. Accepted 9 May 2011 On the importance of intraspecific variability
Distance-Based Functional Diversity Measures and Their Decomposition: A Framework Based on Hill Numbers
 and Their Decomposition: A Framework Based on Hill Numbers Chun-Huo Chiu, Anne Chao* Institute of Statistics, National Tsing Hua University, Hsin-Chu, Taiwan Abstract Hill numbers (or the effective number
and Their Decomposition: A Framework Based on Hill Numbers Chun-Huo Chiu, Anne Chao* Institute of Statistics, National Tsing Hua University, Hsin-Chu, Taiwan Abstract Hill numbers (or the effective number
SPECIAL FEATURE: FUNCTIONAL DIVERSITY Which trait dissimilarity for functional diversity: trait means or trait overlap?
 && (2012) SPECIAL FEATURE: FUNCTIONAL DIVERSITY Which trait dissimilarity for functional diversity: trait means or trait overlap? Francesco de Bello, Carlos P. Carmona, Norman W. H. Mason, Maria-Teresa
&& (2012) SPECIAL FEATURE: FUNCTIONAL DIVERSITY Which trait dissimilarity for functional diversity: trait means or trait overlap? Francesco de Bello, Carlos P. Carmona, Norman W. H. Mason, Maria-Teresa
How to quantify biological diversity: taxonomical, functional and evolutionary aspects. Hanna Tuomisto, University of Turku
 How to quantify biological diversity: taxonomical, functional and evolutionary aspects Hanna Tuomisto, University of Turku Why quantify biological diversity? understanding the structure and function of
How to quantify biological diversity: taxonomical, functional and evolutionary aspects Hanna Tuomisto, University of Turku Why quantify biological diversity? understanding the structure and function of
Package hierdiversity
 Version 0.1 Date 2015-03-11 Package hierdiversity March 20, 2015 Title Hierarchical Multiplicative Partitioning of Complex Phenotypes Author Zachary Marion, James Fordyce, and Benjamin Fitzpatrick Maintainer
Version 0.1 Date 2015-03-11 Package hierdiversity March 20, 2015 Title Hierarchical Multiplicative Partitioning of Complex Phenotypes Author Zachary Marion, James Fordyce, and Benjamin Fitzpatrick Maintainer
LEA: An R package for Landscape and Ecological Association Studies. Olivier Francois Ecole GENOMENV AgroParisTech, Paris, 2016
 LEA: An R package for Landscape and Ecological Association Studies Olivier Francois Ecole GENOMENV AgroParisTech, Paris, 2016 Outline Installing LEA Formatting the data for LEA Basic principles o o Analysis
LEA: An R package for Landscape and Ecological Association Studies Olivier Francois Ecole GENOMENV AgroParisTech, Paris, 2016 Outline Installing LEA Formatting the data for LEA Basic principles o o Analysis
Chapter 2 Exploratory Data Analysis
 Chapter 2 Exploratory Data Analysis 2.1 Objectives Nowadays, most ecological research is done with hypothesis testing and modelling in mind. However, Exploratory Data Analysis (EDA), which uses visualization
Chapter 2 Exploratory Data Analysis 2.1 Objectives Nowadays, most ecological research is done with hypothesis testing and modelling in mind. However, Exploratory Data Analysis (EDA), which uses visualization
Package Select. May 11, 2018
 Package Select May 11, 2018 Title Determines Species Probabilities Based on Functional Traits Version 1.4 The objective of these functions is to derive a species assemblage that satisfies a functional
Package Select May 11, 2018 Title Determines Species Probabilities Based on Functional Traits Version 1.4 The objective of these functions is to derive a species assemblage that satisfies a functional
Research Article An Index for Measuring Functional Diversity in Plant Communities Based on Neural Network Theory
 Applied Mathematics Volume 213, Article ID 3295, 6 pages http://dx.doi.org/1.1155/213/3295 Research Article An Index for Measuring Functional Diversity in Plant Communities Based on Neural Network Theory
Applied Mathematics Volume 213, Article ID 3295, 6 pages http://dx.doi.org/1.1155/213/3295 Research Article An Index for Measuring Functional Diversity in Plant Communities Based on Neural Network Theory
Studying the effect of species dominance on diversity patterns using Hill numbers-based indices
 Studying the effect of species dominance on diversity patterns using Hill numbers-based indices Loïc Chalmandrier Loïc Chalmandrier Diversity pattern analysis November 8th 2017 1 / 14 Introduction Diversity
Studying the effect of species dominance on diversity patterns using Hill numbers-based indices Loïc Chalmandrier Loïc Chalmandrier Diversity pattern analysis November 8th 2017 1 / 14 Introduction Diversity
Ch20_Ecology, community & ecosystems
 Community Ecology Populations of different species living in the same place NICHE The sum of all the different use of abiotic resources in the habitat by s given species what the organism does what is
Community Ecology Populations of different species living in the same place NICHE The sum of all the different use of abiotic resources in the habitat by s given species what the organism does what is
Variable-Specific Entropy Contribution
 Variable-Specific Entropy Contribution Tihomir Asparouhov and Bengt Muthén June 19, 2018 In latent class analysis it is useful to evaluate a measurement instrument in terms of how well it identifies the
Variable-Specific Entropy Contribution Tihomir Asparouhov and Bengt Muthén June 19, 2018 In latent class analysis it is useful to evaluate a measurement instrument in terms of how well it identifies the
Assessing functional diversity in the field methodology matters!
 Functional Ecology 2007 doi: 10.1111/j.1365-2435.2007.01339.x Blackwell Publishing Ltd REVIEW Assessing functional diversity in the field methodology matters! Sandra Lavorel 1,2 *, Karl Grigulis 1,2, Sue
Functional Ecology 2007 doi: 10.1111/j.1365-2435.2007.01339.x Blackwell Publishing Ltd REVIEW Assessing functional diversity in the field methodology matters! Sandra Lavorel 1,2 *, Karl Grigulis 1,2, Sue
Functional diversity in plant communities: Theory and analysis methods
 African Journal of Biotechnology Vol. (5), pp. 04-022, 6 January, 202 Available online at http://www.academicjournals.org/ajb DOI: 0.5897/AJB.322 ISSN 684 535 202 Academic Journals Review Functional diversity
African Journal of Biotechnology Vol. (5), pp. 04-022, 6 January, 202 Available online at http://www.academicjournals.org/ajb DOI: 0.5897/AJB.322 ISSN 684 535 202 Academic Journals Review Functional diversity
Extending the concept of diversity partitioning to characterize phenotypic complexity. Zachary Marion James Fordyce Ben Fitzpatrick
 Extending the concept of diversity partitioning to characterize phenotypic complexity Zachary Marion James Fordyce Ben Fitzpatrick Acknowledgements Fireflies Lynn Faust Raphael De Cock Kathrin Stanger-Hall
Extending the concept of diversity partitioning to characterize phenotypic complexity Zachary Marion James Fordyce Ben Fitzpatrick Acknowledgements Fireflies Lynn Faust Raphael De Cock Kathrin Stanger-Hall
Multivariate Analysis of Ecological Data
 Multivariate Analysis of Ecological Data MICHAEL GREENACRE Professor of Statistics at the Pompeu Fabra University in Barcelona, Spain RAUL PRIMICERIO Associate Professor of Ecology, Evolutionary Biology
Multivariate Analysis of Ecological Data MICHAEL GREENACRE Professor of Statistics at the Pompeu Fabra University in Barcelona, Spain RAUL PRIMICERIO Associate Professor of Ecology, Evolutionary Biology
Metacommunities Spatial Ecology of Communities
 Spatial Ecology of Communities Four perspectives for multiple species Patch dynamics principles of metapopulation models (patchy pops, Levins) Mass effects principles of source-sink and rescue effects
Spatial Ecology of Communities Four perspectives for multiple species Patch dynamics principles of metapopulation models (patchy pops, Levins) Mass effects principles of source-sink and rescue effects
EXAM PRACTICE. 12 questions * 4 categories: Statistics Background Multivariate Statistics Interpret True / False
 EXAM PRACTICE 12 questions * 4 categories: Statistics Background Multivariate Statistics Interpret True / False Stats 1: What is a Hypothesis? A testable assertion about how the world works Hypothesis
EXAM PRACTICE 12 questions * 4 categories: Statistics Background Multivariate Statistics Interpret True / False Stats 1: What is a Hypothesis? A testable assertion about how the world works Hypothesis
Deciphering the Enigma of Undetected Species, Phylogenetic, and Functional Diversity. Based on Good-Turing Theory
 Metadata S1 Deciphering the Enigma of Undetected Species, Phylogenetic, and Functional Diversity Based on Good-Turing Theory Anne Chao, Chun-Huo Chiu, Robert K. Colwell, Luiz Fernando S. Magnago, Robin
Metadata S1 Deciphering the Enigma of Undetected Species, Phylogenetic, and Functional Diversity Based on Good-Turing Theory Anne Chao, Chun-Huo Chiu, Robert K. Colwell, Luiz Fernando S. Magnago, Robin
Package velociraptr. February 15, 2017
 Type Package Title Fossil Analysis Version 1.0 Author Package velociraptr February 15, 2017 Maintainer Andrew A Zaffos Functions for downloading, reshaping, culling, cleaning, and analyzing
Type Package Title Fossil Analysis Version 1.0 Author Package velociraptr February 15, 2017 Maintainer Andrew A Zaffos Functions for downloading, reshaping, culling, cleaning, and analyzing
Fernando A. O. Silveira Universidade Federal de Minas Gerais, Brazil
 Fernando A. O. Silveira Universidade Federal de Minas Gerais, Brazil www.leept.webnode.com Inselbergs are geologically-ancient, nutrientimpoverished granitic and gneiss monoliths that rise sharply above
Fernando A. O. Silveira Universidade Federal de Minas Gerais, Brazil www.leept.webnode.com Inselbergs are geologically-ancient, nutrientimpoverished granitic and gneiss monoliths that rise sharply above
Wavelet methods and null models for spatial pattern analysis
 Wavelet methods and null models for spatial pattern analysis Pavel Dodonov Part of my PhD thesis, by the Federal University of São Carlos (São Carlos, SP, Brazil), supervised by Dr Dalva M. Silva-Matos
Wavelet methods and null models for spatial pattern analysis Pavel Dodonov Part of my PhD thesis, by the Federal University of São Carlos (São Carlos, SP, Brazil), supervised by Dr Dalva M. Silva-Matos
Discriminant Analysis Documentation
 Discriminant Analysis Documentation Release 1 Tim Thatcher May 01, 2016 Contents 1 Installation 3 2 Theory 5 2.1 Linear Discriminant Analysis (LDA).................................. 5 2.2 Quadratic Discriminant
Discriminant Analysis Documentation Release 1 Tim Thatcher May 01, 2016 Contents 1 Installation 3 2 Theory 5 2.1 Linear Discriminant Analysis (LDA).................................. 5 2.2 Quadratic Discriminant
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200 Spring 2014 University of California, Berkeley
 "PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200 Spring 2014 University of California, Berkeley D.D. Ackerly April 16, 2014. Community Ecology and Phylogenetics Readings: Cavender-Bares,
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200 Spring 2014 University of California, Berkeley D.D. Ackerly April 16, 2014. Community Ecology and Phylogenetics Readings: Cavender-Bares,
Ecological Significance of Within- Species Leaf Trait Variability: A Test Using an Island Area Gradient
 Ecological Significance of Within- Species Leaf Trait Variability: A Test Using an Island Area Gradient Bright Kumordzi Faculty of Forest Sciences Department of Forest Ecology and Management Umeå Doctoral
Ecological Significance of Within- Species Leaf Trait Variability: A Test Using an Island Area Gradient Bright Kumordzi Faculty of Forest Sciences Department of Forest Ecology and Management Umeå Doctoral
FDiversity: a software package for the integrated analysis of functional diversity
 Methods in Ecology and Evolution 2011, 2, 233 237 doi: 10.1111/j.2041-210X.2010.00082.x APPLICATION FDiversity: a software package for the integrated analysis of functional diversity Fernando Casanoves
Methods in Ecology and Evolution 2011, 2, 233 237 doi: 10.1111/j.2041-210X.2010.00082.x APPLICATION FDiversity: a software package for the integrated analysis of functional diversity Fernando Casanoves
Transitivity a FORTRAN program for the analysis of bivariate competitive interactions Version 1.1
 Transitivity 1 Transitivity a FORTRAN program for the analysis of bivariate competitive interactions Version 1.1 Werner Ulrich Nicolaus Copernicus University in Toruń Chair of Ecology and Biogeography
Transitivity 1 Transitivity a FORTRAN program for the analysis of bivariate competitive interactions Version 1.1 Werner Ulrich Nicolaus Copernicus University in Toruń Chair of Ecology and Biogeography
Testing CSR theory: a single-species approach with Bellevalia webbiana as a case study
 Testing CSR theory: a single-species approach with Bellevalia webbiana as a case study Giovanni Astuti, Daniela Ciccarelli, Francesco Roma-Marzio, Alberto Trinco, Lorenzo Peruzzi CSR tertiary strategy
Testing CSR theory: a single-species approach with Bellevalia webbiana as a case study Giovanni Astuti, Daniela Ciccarelli, Francesco Roma-Marzio, Alberto Trinco, Lorenzo Peruzzi CSR tertiary strategy
Ecohydrologic mechanisms of Mopane in the Kruger National Park
 Ecohydrologic mechanisms of Mopane in the Kruger National Park photo: AK Knapp Jesse B Nippert & Anthony M Swemmer Predicting the FUTURE Savanna types and distributions are susceptible to climate change
Ecohydrologic mechanisms of Mopane in the Kruger National Park photo: AK Knapp Jesse B Nippert & Anthony M Swemmer Predicting the FUTURE Savanna types and distributions are susceptible to climate change
Linking Traits to Ecosystem Processes. Moira Hough
 Linking Traits to Ecosystem Processes Moira Hough How do organisms impact ecosystems? Long history of study of ecological effects of biodiversity and species composi@on comes out of the diversity stability
Linking Traits to Ecosystem Processes Moira Hough How do organisms impact ecosystems? Long history of study of ecological effects of biodiversity and species composi@on comes out of the diversity stability
Plant Functional Traits Improve Diversity-Based Predictions of Temporal Stability of Grassland Productivity
 Ecology, Evolution and Organismal Biology Publications Ecology, Evolution and Organismal Biology 9-2013 Plant Functional Traits Improve Diversity-Based Predictions of Temporal Stability of Grassland Productivity
Ecology, Evolution and Organismal Biology Publications Ecology, Evolution and Organismal Biology 9-2013 Plant Functional Traits Improve Diversity-Based Predictions of Temporal Stability of Grassland Productivity
Intraspecific variation in traits reduces ability of trait-based models to predict community structure
 28 (2017) 1070 1081 Intraspecific variation in traits reduces ability of trait-based models to predict community structure Quentin D. Read, Jeremiah A. Henning & Nathan J. Sanders Keywords Community model;
28 (2017) 1070 1081 Intraspecific variation in traits reduces ability of trait-based models to predict community structure Quentin D. Read, Jeremiah A. Henning & Nathan J. Sanders Keywords Community model;
Introduction to Occupancy Models. Jan 8, 2016 AEC 501 Nathan J. Hostetter
 Introduction to Occupancy Models Jan 8, 2016 AEC 501 Nathan J. Hostetter njhostet@ncsu.edu 1 Occupancy Abundance often most interesting variable when analyzing a population Occupancy probability that a
Introduction to Occupancy Models Jan 8, 2016 AEC 501 Nathan J. Hostetter njhostet@ncsu.edu 1 Occupancy Abundance often most interesting variable when analyzing a population Occupancy probability that a
Octave. Tutorial. Daniel Lamprecht. March 26, Graz University of Technology. Slides based on previous work by Ingo Holzmann
 Tutorial Graz University of Technology March 26, 2012 Slides based on previous work by Ingo Holzmann Introduction What is? GNU is a high-level interactive language for numerical computations mostly compatible
Tutorial Graz University of Technology March 26, 2012 Slides based on previous work by Ingo Holzmann Introduction What is? GNU is a high-level interactive language for numerical computations mostly compatible
Chapter 5: Evenness, Richness and Diversity
 142 Chapter 5: Deep crevice in a large Inselberg at Bornhardtia near Ironbark 143 Chapter 5 Eveness, Richness and Diversity 5.1 Introduction The distribution of abundances amongst species in communities
142 Chapter 5: Deep crevice in a large Inselberg at Bornhardtia near Ironbark 143 Chapter 5 Eveness, Richness and Diversity 5.1 Introduction The distribution of abundances amongst species in communities
Dissimilarity and transformations. Pierre Legendre Département de sciences biologiques Université de Montréal
 and transformations Pierre Legendre Département de sciences biologiques Université de Montréal http://www.numericalecology.com/ Pierre Legendre 2017 Definitions An association coefficient is a function
and transformations Pierre Legendre Département de sciences biologiques Université de Montréal http://www.numericalecology.com/ Pierre Legendre 2017 Definitions An association coefficient is a function
Norman W.H. Mason, Francesco de Bello, David Mouillot, Sandrine Pavoine & Stéphane Dray
 24 (2013) 794 806 SPECIAL FEATURE: FUNCTIONAL DIVERSITY A guide for using functional diversity indices to reveal changes in assembly processes along ecological gradients Norman W.H. Mason, Francesco de
24 (2013) 794 806 SPECIAL FEATURE: FUNCTIONAL DIVERSITY A guide for using functional diversity indices to reveal changes in assembly processes along ecological gradients Norman W.H. Mason, Francesco de
Towards a consensus for calculating dendrogram-based functional diversity indices
 Oikos 117: 794800, 2008 doi: 10.1111/j.2008.0030-1299.16594.x, # The Authors. Journal compilation # Oikos 2008 Subject Editor: Nicholas Gotelli, Accepted 1 February 2008 Towards a consensus for calculating
Oikos 117: 794800, 2008 doi: 10.1111/j.2008.0030-1299.16594.x, # The Authors. Journal compilation # Oikos 2008 Subject Editor: Nicholas Gotelli, Accepted 1 February 2008 Towards a consensus for calculating
Consequences of biodiversity loss diverge from expectation due to. Ocean and Earth Science, National Oceanography Centre Southampton,
 Supplementary Material Consequences of biodiversity loss diverge from expectation due to post-extinction compensatory responses Authors: Matthias S. Thomsen 1*, Clement Garcia 2, Stefan G. Bolam 2, Ruth
Supplementary Material Consequences of biodiversity loss diverge from expectation due to post-extinction compensatory responses Authors: Matthias S. Thomsen 1*, Clement Garcia 2, Stefan G. Bolam 2, Ruth
What is a meta-analysis? How is a meta-analysis conducted? Model Selection Approaches to Inference. Meta-analysis. Combining Data
 Combining Data IB/NRES 509 Statistical Modeling What is a? A quantitative synthesis of previous research Studies as individual observations, weighted by n, σ 2, quality, etc. Can combine heterogeneous
Combining Data IB/NRES 509 Statistical Modeling What is a? A quantitative synthesis of previous research Studies as individual observations, weighted by n, σ 2, quality, etc. Can combine heterogeneous
Functional Diversity. By Morgan Davies and Emily Smith
 Functional Diversity By Morgan Davies and Emily Smith Outline Introduction to biodiversity and functional diversity How do we measure functional diversity Why do we care about functional diversity Applications
Functional Diversity By Morgan Davies and Emily Smith Outline Introduction to biodiversity and functional diversity How do we measure functional diversity Why do we care about functional diversity Applications
Aboveground resilience to species loss but belowground resistance to nitrogen addition in a montane plant community
 Journal of Plant Ecology VOLUME 11, NUMBER 3, PAGES 351 363 June 2018 doi: 10.1093/jpe/rtx015 Advance Access publication 24 February 2017 available online at academic.oup.com/jpe Abstract Aims Decades
Journal of Plant Ecology VOLUME 11, NUMBER 3, PAGES 351 363 June 2018 doi: 10.1093/jpe/rtx015 Advance Access publication 24 February 2017 available online at academic.oup.com/jpe Abstract Aims Decades
Using the EartH2Observe data portal to analyse drought indicators. Lesson 4: Using Python Notebook to access and process data
 Using the EartH2Observe data portal to analyse drought indicators Lesson 4: Using Python Notebook to access and process data Preface In this fourth lesson you will again work with the Water Cycle Integrator
Using the EartH2Observe data portal to analyse drought indicators Lesson 4: Using Python Notebook to access and process data Preface In this fourth lesson you will again work with the Water Cycle Integrator
Spatial non-stationarity, anisotropy and scale: The interactive visualisation of spatial turnover
 19th International Congress on Modelling and Simulation, Perth, Australia, 12 16 December 2011 http://mssanz.org.au/modsim2011 Spatial non-stationarity, anisotropy and scale: The interactive visualisation
19th International Congress on Modelling and Simulation, Perth, Australia, 12 16 December 2011 http://mssanz.org.au/modsim2011 Spatial non-stationarity, anisotropy and scale: The interactive visualisation
What is the range of a taxon? A scaling problem at three levels: Spa9al scale Phylogene9c depth Time
 What is the range of a taxon? A scaling problem at three levels: Spa9al scale Phylogene9c depth Time 1 5 0.25 0.15 5 0.05 0.05 0.10 2 0.10 0.10 0.20 4 Reminder of what a range-weighted tree is Actual Tree
What is the range of a taxon? A scaling problem at three levels: Spa9al scale Phylogene9c depth Time 1 5 0.25 0.15 5 0.05 0.05 0.10 2 0.10 0.10 0.20 4 Reminder of what a range-weighted tree is Actual Tree
Extremes analysis: the. ETCCDI twopronged
 Manola Brunet University Rovira i Virgili, Tarragona, Catalonia Extremes analysis: the Title ETCCDI twopronged approach 5 December 2017 Fourth Session of the ETCCDI, Victoria, Feb 2011 The role of the
Manola Brunet University Rovira i Virgili, Tarragona, Catalonia Extremes analysis: the Title ETCCDI twopronged approach 5 December 2017 Fourth Session of the ETCCDI, Victoria, Feb 2011 The role of the
Appendix S1 Replacement, richness difference and nestedness indices
 Appendix to: Legendre, P. (2014) Interpreting the replacement and richness difference components of beta diversity. Global Ecology and Biogeography, 23, 1324-1334. Appendix S1 Replacement, richness difference
Appendix to: Legendre, P. (2014) Interpreting the replacement and richness difference components of beta diversity. Global Ecology and Biogeography, 23, 1324-1334. Appendix S1 Replacement, richness difference
1 Soil Factors Affecting Nutrient Bioavailability... 1 N.B. Comerford
 Contents 1 Soil Factors Affecting Nutrient Bioavailability........ 1 N.B. Comerford 1.1 Introduction........................... 1 1.2 Release of Nutrients from the Soil Solid Phase........ 2 1.3 Nutrient
Contents 1 Soil Factors Affecting Nutrient Bioavailability........ 1 N.B. Comerford 1.1 Introduction........................... 1 1.2 Release of Nutrients from the Soil Solid Phase........ 2 1.3 Nutrient
MITOCW MITRES2_002S10nonlinear_lec05_300k-mp4
 MITOCW MITRES2_002S10nonlinear_lec05_300k-mp4 The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources
MITOCW MITRES2_002S10nonlinear_lec05_300k-mp4 The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources
Yakın Doğu Üniversitesi Mimarlık Fakültesi Peyzaj Mimarlığı Bölümü. PM 317 Human and Environment Assoc. Prof. Dr. Salih GÜCEL
 Yakın Doğu Üniversitesi Mimarlık Fakültesi Peyzaj Mimarlığı Bölümü PM 317 Human and Environment Assoc. Prof. Dr. Salih GÜCEL Ecology & Ecosystems Principles of Ecology Ecology is the study of the interactions
Yakın Doğu Üniversitesi Mimarlık Fakültesi Peyzaj Mimarlığı Bölümü PM 317 Human and Environment Assoc. Prof. Dr. Salih GÜCEL Ecology & Ecosystems Principles of Ecology Ecology is the study of the interactions
Changes in local-scale intraspecific trait variability of dominant species across contrasting island ecosystems
 Changes in local-scale intraspecific trait variability of dominant species across contrasting island ecosystems BRIGHT B. KUMORDZI, MARIE-CHARLOTTE NILSSON, MICHAEL J. GUNDALE, AND DAVID A. WARDLE Department
Changes in local-scale intraspecific trait variability of dominant species across contrasting island ecosystems BRIGHT B. KUMORDZI, MARIE-CHARLOTTE NILSSON, MICHAEL J. GUNDALE, AND DAVID A. WARDLE Department
Invertibility of random matrices
 University of Michigan February 2011, Princeton University Origins of Random Matrix Theory Statistics (Wishart matrices) PCA of a multivariate Gaussian distribution. [Gaël Varoquaux s blog gael-varoquaux.info]
University of Michigan February 2011, Princeton University Origins of Random Matrix Theory Statistics (Wishart matrices) PCA of a multivariate Gaussian distribution. [Gaël Varoquaux s blog gael-varoquaux.info]
Individual-level trait diversity concepts and indices to comprehensively describe community change in multidimensional trait space
 Functional Ecology 2016, 30, 808818 doi: 10.1111/1365-2435.12551 Individual-level trait diversity concepts and indices to comprehensively describe community change in multidimensional trait space Simone
Functional Ecology 2016, 30, 808818 doi: 10.1111/1365-2435.12551 Individual-level trait diversity concepts and indices to comprehensively describe community change in multidimensional trait space Simone
Domain decomposition on different levels of the Jacobi-Davidson method
 hapter 5 Domain decomposition on different levels of the Jacobi-Davidson method Abstract Most computational work of Jacobi-Davidson [46], an iterative method suitable for computing solutions of large dimensional
hapter 5 Domain decomposition on different levels of the Jacobi-Davidson method Abstract Most computational work of Jacobi-Davidson [46], an iterative method suitable for computing solutions of large dimensional
The effects of grazing on foliar trait diversity and niche differentiation in Tibetan alpine meadows
 The effects of grazing on foliar trait diversity and niche differentiation in Tibetan alpine meadows KECHANG NIU, 1,2,3, JULIE MESSIER, 3,4 JIN-SHENG HE, 2 AND MARTIN J. LECHOWICZ 3 1 Department of Biology,
The effects of grazing on foliar trait diversity and niche differentiation in Tibetan alpine meadows KECHANG NIU, 1,2,3, JULIE MESSIER, 3,4 JIN-SHENG HE, 2 AND MARTIN J. LECHOWICZ 3 1 Department of Biology,
diversity(datamatrix, index= shannon, base=exp(1))
 Tutorial 11: Diversity, Indicator Species Analysis, Cluster Analysis Calculating Diversity Indices The vegan package contains the command diversity() for calculating Shannon and Simpson diversity indices.
Tutorial 11: Diversity, Indicator Species Analysis, Cluster Analysis Calculating Diversity Indices The vegan package contains the command diversity() for calculating Shannon and Simpson diversity indices.
Science 9 - Unit A Review Sheet
 Science 9 - Unit A Review Sheet Learning Outcomes Can you? describe the relative abundance of species on Earth and in different environments describe examples of variation among species and within species
Science 9 - Unit A Review Sheet Learning Outcomes Can you? describe the relative abundance of species on Earth and in different environments describe examples of variation among species and within species
The Solution to Assignment 6
 The Solution to Assignment 6 Problem 1: Use the 2-fold cross-validation to evaluate the Decision Tree Model for trees up to 2 levels deep (that is, the maximum path length from the root to the leaves is
The Solution to Assignment 6 Problem 1: Use the 2-fold cross-validation to evaluate the Decision Tree Model for trees up to 2 levels deep (that is, the maximum path length from the root to the leaves is
Introduction to Independent Component Analysis. Jingmei Lu and Xixi Lu. Abstract
 Final Project 2//25 Introduction to Independent Component Analysis Abstract Independent Component Analysis (ICA) can be used to solve blind signal separation problem. In this article, we introduce definition
Final Project 2//25 Introduction to Independent Component Analysis Abstract Independent Component Analysis (ICA) can be used to solve blind signal separation problem. In this article, we introduce definition
Evolution of phenotypic traits
 Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus.
 PCI GEOMATICS GEOMATICA QUICKSTART 1. Introduction This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus. All data used in
PCI GEOMATICS GEOMATICA QUICKSTART 1. Introduction This tutorial is intended to familiarize you with the Geomatica Toolbar and describe the basics of viewing data using Geomatica Focus. All data used in
Lecture 08: Standard methods. Bruce Walsh lecture notes Tucson Winter Institute 9-11 Jan 2013
 Lecture 08: G x E: Genotype-byenvironment interactions: Standard methods Bruce Walsh lecture notes Tucson Winter Institute 9-11 Jan 2013 1 G x E Introduction to G x E Basics of G x E G x E is a correlated
Lecture 08: G x E: Genotype-byenvironment interactions: Standard methods Bruce Walsh lecture notes Tucson Winter Institute 9-11 Jan 2013 1 G x E Introduction to G x E Basics of G x E G x E is a correlated
G E INTERACTION USING JMP: AN OVERVIEW
 G E INTERACTION USING JMP: AN OVERVIEW Sukanta Dash I.A.S.R.I., Library Avenue, New Delhi-110012 sukanta@iasri.res.in 1. Introduction Genotype Environment interaction (G E) is a common phenomenon in agricultural
G E INTERACTION USING JMP: AN OVERVIEW Sukanta Dash I.A.S.R.I., Library Avenue, New Delhi-110012 sukanta@iasri.res.in 1. Introduction Genotype Environment interaction (G E) is a common phenomenon in agricultural
What determines: 1) Species distributions? 2) Species diversity? Patterns and processes
 Species diversity What determines: 1) Species distributions? 2) Species diversity? Patterns and processes At least 120 different (overlapping) hypotheses explaining species richness... We are going to
Species diversity What determines: 1) Species distributions? 2) Species diversity? Patterns and processes At least 120 different (overlapping) hypotheses explaining species richness... We are going to
Functional diversity measures: an overview of their redundancy and their ability to discriminate community assembly rules
 Functional Ecology 2010, 24, 867 876 doi: 10.1111/j.1365-2435.2010.01695.x Functional diversity measures: an overview of their redundancy and their ability to discriminate community assembly rules Maud
Functional Ecology 2010, 24, 867 876 doi: 10.1111/j.1365-2435.2010.01695.x Functional diversity measures: an overview of their redundancy and their ability to discriminate community assembly rules Maud
Ecosystems. 1. Population Interactions 2. Energy Flow 3. Material Cycle
 Ecosystems 1. Population Interactions 2. Energy Flow 3. Material Cycle The deep sea was once thought to have few forms of life because of the darkness (no photosynthesis) and tremendous pressures. But
Ecosystems 1. Population Interactions 2. Energy Flow 3. Material Cycle The deep sea was once thought to have few forms of life because of the darkness (no photosynthesis) and tremendous pressures. But
Wolfgang Karl Härdle Leopold Simar. Applied Multivariate. Statistical Analysis. Fourth Edition. ö Springer
 Wolfgang Karl Härdle Leopold Simar Applied Multivariate Statistical Analysis Fourth Edition ö Springer Contents Part I Descriptive Techniques 1 Comparison of Batches 3 1.1 Boxplots 4 1.2 Histograms 11
Wolfgang Karl Härdle Leopold Simar Applied Multivariate Statistical Analysis Fourth Edition ö Springer Contents Part I Descriptive Techniques 1 Comparison of Batches 3 1.1 Boxplots 4 1.2 Histograms 11
Overview of Chapter 5
 Chapter 5 Ecosystems and Living Organisms Overview of Chapter 5 Evolution Natural Selection Biological Communities Symbiosis Predation & Competition Community Development Succession Evolution The cumulative
Chapter 5 Ecosystems and Living Organisms Overview of Chapter 5 Evolution Natural Selection Biological Communities Symbiosis Predation & Competition Community Development Succession Evolution The cumulative
Community Ecology Bio 147/247 Species Richness 3: Diversity& Abundance Deeper Meanings of Biodiversity Speci es and Functional Groups
 Community Ecology Bio 147/247 Species Richness 3: Diversity& Abundance Deeper Meanings of Biodiversity Speci es and Functional Groups The main Qs for today are: 1. How many species are there in a community?
Community Ecology Bio 147/247 Species Richness 3: Diversity& Abundance Deeper Meanings of Biodiversity Speci es and Functional Groups The main Qs for today are: 1. How many species are there in a community?
Valley Central School District 944 State Route 17K Montgomery, NY Telephone Number: (845) ext Fax Number: (845)
 Valley Central School District 944 State Route 17K Montgomery, NY 12549 Telephone Number: (845)457-2400 ext. 18121 Fax Number: (845)457-4254 Advance Placement Biology Presented to the Board of Education
Valley Central School District 944 State Route 17K Montgomery, NY 12549 Telephone Number: (845)457-2400 ext. 18121 Fax Number: (845)457-4254 Advance Placement Biology Presented to the Board of Education
Package HierDpart. February 13, 2019
 Version 0.3.5 Date 2019-02-10 Package HierDpart February 13, 2019 Title Partitioning Hierarchical Diversity and Differentiation Across Metrics and Scales, from Genes to Ecosystems Miscellaneous R functions
Version 0.3.5 Date 2019-02-10 Package HierDpart February 13, 2019 Title Partitioning Hierarchical Diversity and Differentiation Across Metrics and Scales, from Genes to Ecosystems Miscellaneous R functions
An introduction to the picante package
 An introduction to the picante package Steven Kembel (steve.kembel@gmail.com) April 2010 Contents 1 Installing picante 1 2 Data formats in picante 2 2.1 Phylogenies................................ 2 2.2
An introduction to the picante package Steven Kembel (steve.kembel@gmail.com) April 2010 Contents 1 Installing picante 1 2 Data formats in picante 2 2.1 Phylogenies................................ 2 2.2
Evidence for Competition
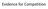 Evidence for Competition Population growth in laboratory experiments carried out by the Russian scientist Gause on growth rates in two different yeast species Each of the species has the same food e.g.,
Evidence for Competition Population growth in laboratory experiments carried out by the Russian scientist Gause on growth rates in two different yeast species Each of the species has the same food e.g.,
Current topics in clonal plants research: editorial
 Preslia 83: 275 279, 2011 275 Current topics in clonal plants research: editorial Současné trendy ve výzkumu klonálních rostlin Jitka K l i m e š o v á 1 & Petr P y š e k 2, 3 1 Institute of Botany, Academy
Preslia 83: 275 279, 2011 275 Current topics in clonal plants research: editorial Současné trendy ve výzkumu klonálních rostlin Jitka K l i m e š o v á 1 & Petr P y š e k 2, 3 1 Institute of Botany, Academy
Introduction to Computational Neuroscience
 CSE2330 Introduction to Computational Neuroscience Basic computational tools and concepts Tutorial 1 Duration: two weeks 1.1 About this tutorial The objective of this tutorial is to introduce you to: the
CSE2330 Introduction to Computational Neuroscience Basic computational tools and concepts Tutorial 1 Duration: two weeks 1.1 About this tutorial The objective of this tutorial is to introduce you to: the
From diversity indices to community assembly processes: a test with simulated data
 Ecography 34: 001 013, 2011 doi: 10.1111/j.1600-0587.2011.07259.x 2011 The Authors. Journal compilation 2011 Nordic Society Oikos Subject Editor: Alexandre Diniz-Filho. Accepted 13 September 2011 From
Ecography 34: 001 013, 2011 doi: 10.1111/j.1600-0587.2011.07259.x 2011 The Authors. Journal compilation 2011 Nordic Society Oikos Subject Editor: Alexandre Diniz-Filho. Accepted 13 September 2011 From
LECTURE 07: CROP GROWTH ANALYSIS
 http://smtom.lecture.ub.ac.id/ Password: https://syukur16tom.wordpress.com/ Password: LECTURE 07: CROP GROWTH ANALYSIS Leaf area was the main factor determining differences in yield in several crops. Watson
http://smtom.lecture.ub.ac.id/ Password: https://syukur16tom.wordpress.com/ Password: LECTURE 07: CROP GROWTH ANALYSIS Leaf area was the main factor determining differences in yield in several crops. Watson
Dynamic Global Vegetation Models. Rosie Fisher Terrestrial Sciences Section, NCAR
 Dynamic Global Vegetation Models Rosie Fisher Terrestrial Sciences Section, NCAR What is the D in DGVM? Recruitment Assimilation Growth Competition Movement of vegetation in space predicted by model Mortality
Dynamic Global Vegetation Models Rosie Fisher Terrestrial Sciences Section, NCAR What is the D in DGVM? Recruitment Assimilation Growth Competition Movement of vegetation in space predicted by model Mortality
RRQR Factorization Linux and Windows MEX-Files for MATLAB
 Documentation RRQR Factorization Linux and Windows MEX-Files for MATLAB March 29, 2007 1 Contents of the distribution file The distribution file contains the following files: rrqrgate.dll: the Windows-MEX-File;
Documentation RRQR Factorization Linux and Windows MEX-Files for MATLAB March 29, 2007 1 Contents of the distribution file The distribution file contains the following files: rrqrgate.dll: the Windows-MEX-File;
Intraspecific trait variation and community assembly in old-field grasslands
 Syracuse University SURFACE Dissertations - ALL SURFACE June 2014 Intraspecific trait variation and community assembly in old-field grasslands Andrew Siefert Syracuse University Follow this and additional
Syracuse University SURFACE Dissertations - ALL SURFACE June 2014 Intraspecific trait variation and community assembly in old-field grasslands Andrew Siefert Syracuse University Follow this and additional
Computer Science & Engineering 423/823 Design and Analysis of Algorithms
 Computer Science & Engineering 423/823 Design and Analysis of Algorithms Lecture 03 Dynamic Programming (Chapter 15) Stephen Scott and Vinodchandran N. Variyam sscott@cse.unl.edu 1/44 Introduction Dynamic
Computer Science & Engineering 423/823 Design and Analysis of Algorithms Lecture 03 Dynamic Programming (Chapter 15) Stephen Scott and Vinodchandran N. Variyam sscott@cse.unl.edu 1/44 Introduction Dynamic
Stability and complexity in model ecosystems
 Stability and complexity in model ecosystems Level 2 module in Modelling course in population and evolutionary biology (701-1418-00) Module author: Sebastian Bonhoeffer Course director: Sebastian Bonhoeffer
Stability and complexity in model ecosystems Level 2 module in Modelling course in population and evolutionary biology (701-1418-00) Module author: Sebastian Bonhoeffer Course director: Sebastian Bonhoeffer
Flexible phenotype simulation with PhenotypeSimulator Hannah Meyer
 Flexible phenotype simulation with PhenotypeSimulator Hannah Meyer 2018-03-01 Contents Introduction 1 Work-flow 2 Examples 2 Example 1: Creating a phenotype composed of population structure and observational
Flexible phenotype simulation with PhenotypeSimulator Hannah Meyer 2018-03-01 Contents Introduction 1 Work-flow 2 Examples 2 Example 1: Creating a phenotype composed of population structure and observational
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 7 Approximate
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 7 Approximate
Chapter 11 Canonical analysis
 Chapter 11 Canonical analysis 11.0 Principles of canonical analysis Canonical analysis is the simultaneous analysis of two, or possibly several data tables. Canonical analyses allow ecologists to perform
Chapter 11 Canonical analysis 11.0 Principles of canonical analysis Canonical analysis is the simultaneous analysis of two, or possibly several data tables. Canonical analyses allow ecologists to perform
User Guide for Hermir version 0.9: Toolbox for Synthesis of Multivariate Stationary Gaussian and non-gaussian Series
 User Guide for Hermir version 0.9: Toolbox for Synthesis of Multivariate Stationary Gaussian and non-gaussian Series Hannes Helgason, Vladas Pipiras, and Patrice Abry June 2, 2011 Contents 1 Organization
User Guide for Hermir version 0.9: Toolbox for Synthesis of Multivariate Stationary Gaussian and non-gaussian Series Hannes Helgason, Vladas Pipiras, and Patrice Abry June 2, 2011 Contents 1 Organization
BAT Biodiversity Assessment Tools, an R package for the measurement and estimation of alpha and beta taxon, phylogenetic and functional diversity
 Methods in Ecology and Evolution 2015, 6, 232 236 doi: 10.1111/2041-210X.12310 APPLICATION BAT Biodiversity Assessment Tools, an R package for the measurement and estimation of alpha and beta taxon, phylogenetic
Methods in Ecology and Evolution 2015, 6, 232 236 doi: 10.1111/2041-210X.12310 APPLICATION BAT Biodiversity Assessment Tools, an R package for the measurement and estimation of alpha and beta taxon, phylogenetic
Linking species-compositional dissimilarities and environmental data for biodiversity assessment
 Linking species-compositional dissimilarities and environmental data for biodiversity assessment D. P. Faith, S. Ferrier Australian Museum, 6 College St., Sydney, N.S.W. 2010, Australia; N.S.W. National
Linking species-compositional dissimilarities and environmental data for biodiversity assessment D. P. Faith, S. Ferrier Australian Museum, 6 College St., Sydney, N.S.W. 2010, Australia; N.S.W. National
Variations in species and functional plant diversity along climatic and grazing gradients
 ECOGRAPHY 29: 801810, 2006 Variations in species and functional plant diversity along climatic and grazing gradients Francesco de Bello, Jan Lepš and Maria-Teresa Sebastià de Bello, F., Lepš, J. and Sebastià,
ECOGRAPHY 29: 801810, 2006 Variations in species and functional plant diversity along climatic and grazing gradients Francesco de Bello, Jan Lepš and Maria-Teresa Sebastià de Bello, F., Lepš, J. and Sebastià,
MULTIVARIATE ANALYSIS OF VARIANCE
 MULTIVARIATE ANALYSIS OF VARIANCE RAJENDER PARSAD AND L.M. BHAR Indian Agricultural Statistics Research Institute Library Avenue, New Delhi - 0 0 lmb@iasri.res.in. Introduction In many agricultural experiments,
MULTIVARIATE ANALYSIS OF VARIANCE RAJENDER PARSAD AND L.M. BHAR Indian Agricultural Statistics Research Institute Library Avenue, New Delhi - 0 0 lmb@iasri.res.in. Introduction In many agricultural experiments,
Topic 4: Orthogonal Contrasts
 Topic 4: Orthogonal Contrasts ANOVA is a useful and powerful tool to compare several treatment means. In comparing t treatments, the null hypothesis tested is that the t true means are all equal (H 0 :
Topic 4: Orthogonal Contrasts ANOVA is a useful and powerful tool to compare several treatment means. In comparing t treatments, the null hypothesis tested is that the t true means are all equal (H 0 :
Gaussian Models
 Gaussian Models ddebarr@uw.edu 2016-04-28 Agenda Introduction Gaussian Discriminant Analysis Inference Linear Gaussian Systems The Wishart Distribution Inferring Parameters Introduction Gaussian Density
Gaussian Models ddebarr@uw.edu 2016-04-28 Agenda Introduction Gaussian Discriminant Analysis Inference Linear Gaussian Systems The Wishart Distribution Inferring Parameters Introduction Gaussian Density
Package MBI. February 19, 2015
 Type Package Package MBI February 19, 2015 Title (M)ultiple-site (B)iodiversity (I)ndices Calculator Version 1.0 Date 2012-10-17 Author Youhua Chen Maintainer Over 20 multiple-site diversity indices can
Type Package Package MBI February 19, 2015 Title (M)ultiple-site (B)iodiversity (I)ndices Calculator Version 1.0 Date 2012-10-17 Author Youhua Chen Maintainer Over 20 multiple-site diversity indices can
2/7/2018. Strata. Strata
 The strata option allows you to control how permutations are done. Specifically, to constrain permutations. Why would you want to do this? In this dataset, there are clear differences in area (A vs. B),
The strata option allows you to control how permutations are done. Specifically, to constrain permutations. Why would you want to do this? In this dataset, there are clear differences in area (A vs. B),
Role of mycorrhizal fungi in belowground C and N cycling
 Role of mycorrhizal fungi in belowground C and N cycling Doc. Jussi Heinonsalo Department of Forest Sciences, University of Helsinki Finnish Meteorological Institute Finland The aim and learning goals
Role of mycorrhizal fungi in belowground C and N cycling Doc. Jussi Heinonsalo Department of Forest Sciences, University of Helsinki Finnish Meteorological Institute Finland The aim and learning goals
A Statistical Distance Approach to Dissimilarities in Ecological Data
 Clemson University TigerPrints All Dissertations Dissertations 5-2015 A Statistical Distance Approach to Dissimilarities in Ecological Data Dominique Jerrod Morgan Clemson University Follow this and additional
Clemson University TigerPrints All Dissertations Dissertations 5-2015 A Statistical Distance Approach to Dissimilarities in Ecological Data Dominique Jerrod Morgan Clemson University Follow this and additional
N, P and O 3 -responses of subalpine plants and their
 Federal Department of Economic Affairs FDEA Agroscope Reckenholz-Tänikon Research Station ART N, P and O 3 -responses of subalpine plants and their mycorrhiza Verena Blanke, Matthias Volk, Seraina Bassin,
Federal Department of Economic Affairs FDEA Agroscope Reckenholz-Tänikon Research Station ART N, P and O 3 -responses of subalpine plants and their mycorrhiza Verena Blanke, Matthias Volk, Seraina Bassin,
Estimating Corn Lily Abundance in Bear Trap Meadow, July 2004
 Estimating Corn Lily Abundance in Bear Trap Meadow, July 2004 Mark Contois, Jameson Cahill, Nacha Chavez, Courtney Cacace, Gail Wechsler, Mariela Pauli, and Ellen Kosman Faculty advisor: Prof. Ed Connor
Estimating Corn Lily Abundance in Bear Trap Meadow, July 2004 Mark Contois, Jameson Cahill, Nacha Chavez, Courtney Cacace, Gail Wechsler, Mariela Pauli, and Ellen Kosman Faculty advisor: Prof. Ed Connor
An Introductory Course in Computational Neuroscience
 An Introductory Course in Computational Neuroscience Contents Series Foreword Acknowledgments Preface 1 Preliminary Material 1.1. Introduction 1.1.1 The Cell, the Circuit, and the Brain 1.1.2 Physics of
An Introductory Course in Computational Neuroscience Contents Series Foreword Acknowledgments Preface 1 Preliminary Material 1.1. Introduction 1.1.1 The Cell, the Circuit, and the Brain 1.1.2 Physics of
