Analytic Computation of the Expectation of the Linkage Disequilibrium Coefficient r 2
|
|
|
- Emmeline Burke
- 5 years ago
- Views:
Transcription
1 Analytic Computation of the Expectation of the Linkage Disequilibrium Coefficient r 2 Yun S. Song a and Jun S. Song b a Department of Computer Science, University of California, Davis, CA 95616, USA b Department of Biostatistics and Computational Biology, Dana-Farber Cancer Institute, Harvard School of Public Health, Boston, MA 02115, USA addresses: yssong@cs.ucdavis.edu (Y.S. Song), jssong@jimmy.harvard.edu (J.S. Song) Corresponding Author: Yun S. Song Department of Computer Science University of California at Davis 2063 Kemper Hall One Shields Avenue Davis, CA U.S.A. yssong@cs.ucdavis.edu Phone: Fax: To appear in Theoretical Population Biology 1
2 Abstract The squared correlation coefficient r 2 (sometimes denoted 2 ) is a measure of linkage disequilibrium that is widely used, but computing its expectation E[r 2 ] in the population has remained an intriguing open problem. The expectation E[r 2 ] is often approximated by the standard linkage deviation σd 2, which is a ratio of two expectations amenable to analytic computation. In this paper, a method of computing the population-wide E[r 2 ] is introduced for a model with recurrent mutation, genetic drift and recombination. The approach is algebraic and is based on the diffusion process approximation. In the limit as the population-scaled recombination rate ρ approaches, it is shown rigorously that the asymptotic behavior of E[r 2 ] is given by 1/ρ + O(ρ 2 ), which, incidentally, is the same as that of σ 2 d. A computer software that computes E[r2 ] numerically is available upon request. Keywords: Linkage disequilibrium, squared correlation coefficient, expectation, diffusion approximation, recurrent mutation, genetic drift, recombination 2
3 1 Introduction Linkage disequilibrium (LD), which characterizes the statistical nonindependence of alleles at different loci, depends on population history and structure, and on various evolutionary forces such as recombination, mutation and natural selection see (Hudson, 2001a) for a general review of LD, and (Pritchard and Przeworski, 2001; International HapMap Consortium, 2005) for studies of LD in the human genome. A question that is of great interest is how one can use LD to learn about the aforementioned factors that influence its extent and patterns. Several different, albeit related, measures of LD between a pair of loci have been proposed. A commonly used measure is r 2 (defined below), which is equal to the square of the correlation coefficient between the alleles at two loci (Hill and Robertson, 1968) and is related to the power of association in gene mapping (see Pritchard and Przeworski 2001 for an explanation of this relationship). In this paper, we consider a diallelic two-locus model for a single panmictic diploid population subject to recurrent mutation and genetic drift. This model is the same as that considered by Ohta and Kimura (1969b). The effective population size, denoted N e, is assumed to remain constant in time. We use A and B to denote the two loci, with allele types A 0, A 1 and B 0, B 1, respectively. The marginal allele frequencies of A 0 and B 0 are respectively denoted by p and q, and the LD measure r 2 is defined as r 2 := D 2 p(1 p)q(1 q), (1) where D = f 00 pq, with f 00 being the frequency of the gametic type A 0 B 0. (Sometimes 2 is used to denote the above LD measure.) We consider the following symmetric reversible mutation model: A 0 u u A 1 and B 0 B 1, u u where u is the mutation rate per generation. The recombination rate between the two loci per generation is denoted by c, and the population-scaled mutation and recombination rates are denoted by θ := 8N e u and ρ := 4N e c, respectively. In his seminal work, Golding (1984) derived a system of recursion relations satisfied by the probabilities of sample configurations, to study the sampling distribution of LD under an infinitelymany-alleles model. This approach was further studied by Ethier and Griffiths (1990), who constructed a two-locus urn model for the distribution of sample configurations. A closed-form solution to the system of recursion relations is not known, but accurate numerical solutions can be obtained for moderate sample sizes (say, up to about 40. See Hudson 2001a). For larger sample sizes, numerically solving the recursion relations becomes intractable, but coalescent simulations can be employed to study the sampling distribution and sample estimated expectations of LD, as done by Hudson (1985, 2001b). A further study of the sampling properties of LD was carried out by Hill and Weir (1994), who used a general forward simulation approach that can easily incorporate 3
4 biologically important features such as selection. In our work, we do not consider the sampling distribution or other sampling properties of LD, but focus on the population-wide expectation of r 2 at equilibrium. (Roughly, the case we consider corresponds to having a very large sample size.) In what follows, p, q, and D denote population quantities. Computing the population expectation E[r 2 ] of the squared correlation coefficient r 2 is a difficult problem, and no analytic approach has been suggested so far. In contrast, as shown by Hill and Robertson (1968) and by Ohta and Kimura (1969a,b), the ratio E[D 2 ]/E[p(1 p)q(1 q)] of two expectations called the standard linkage deviation and often denoted σd 2 is much easier to tackle and closed-form formulae can be obtained for various models. Furthermore, under the neutral model, σd 2 admits a nice genealogical interpretation in terms of covariances in coalescence times (McVean, 2002). The expectation of ratios and the ratio of expectations are, however, of course not the same. Indeed, a previous simulation-based study, in the context of an infinitelymany-alleles model, has shown that σ 2 d may overestimate E[r2 ] by a substantial amount (Maruyama, 1982), although σd 2 is a reasonably good approximation of the sample estimated expectation of r2 conditioned on minor allele frequencies being above 5% (Hudson, 1985). In this paper, we readdress this classic issue and develop an algebraic method of computing the expectation E[r 2 ]. As in the work of Ohta and Kimura (1969a,b), our method is formulated in the context of the diffusion process approximation, which is continuous in both time and space. Diffusion theory can be used to generate useful linear equations satisfied by certain expectations at stationarity, a state in which the effects of mutation, genetic drift, and recombination are in balance. The general idea behind our approach is to express E[r 2 ] in terms of expectations that can be computed by solving appropriate systems of linear equations arising from diffusion theory. We have written a C program, called ER2, that solves the required systems of linear equations numerically. ER2 is available upon request. For given values of θ and ρ, ER2 can produce numerical estimates of E[r 2 ]. We remark that the accuracy of the estimates depends on the chosen truncation level l max, described later in the text; l max = 700 gives very accurate answers, with the running time of a few minutes on a laptop. Our study shows that E[r 2 ] and σd 2 for the assumed model are quite close for large θ (say, θ > 4). For small θ (say, θ 1), however, σd 2 is larger than E[r2 ] by a substantial amount, in some cases by tens of times. If no restriction is imposed on segregation at the two loci, estimates from coalescent simulations are much closer to our theoretical computation of E[r 2 ] than they are to σ 2 d. When conditioned on segregation at both loci, however, sample estimated average r 2 increases substantially for small θ, thus resembling the behavior of σ 2 d. Obtaining a general closed-form formula for E[r 2 ] still remains an open problem, but relevant closed-form expressions can be obtained in the limit ρ. Using our approach, we rigorously show that the large ρ behavior of E[r 2 ] is given by 1/ρ + O(ρ 2 ). Incidentally, this asymptotic behavior of E[r 2 ] is the same as that of σd 2 found by Ohta and Kimura (1969a,b). The organization of this paper is as follows. In Section 2, we describe our method of com- 4
5 puting E[r 2 ] in the context of the diffusion process approximation. In Section 3, we compare our computation of E[r 2 ] with σd 2 and with results from coalescent simulations. The aforementioned exact asymptotic behavior of E[r 2 ] in the large ρ limit is discussed in Section 4. In Section 5, we consider a discrete version of the assumed model and show how combinatorial techniques may be employed to compute certain expectations exactly; such computations are useful for studying the accuracy of the diffusion approximation approach. We conclude in Section 6 with an outlook on future direction. 2 Diffusion Approximation As in (Ohta and Kimura, 1969a,b), our approach is based on diffusion approximation. Being continuous in both time and space, diffusion processes possess many nice properties not shared by discrete processes. In particular, associated to a diffusion process is a fundamental differential operator (i.e., the generator) that has a wide range of applications. As Ohta and Kimura (1969b) have shown, diffusion approximation is a powerful technique that can be used to compute certain expectations at stationarity with surprisingly little effort. In this section, we extend the work of Ohta and Kimura (1969b) to compute the expectation of r Diffusion generator Let L denote the generator of a diffusion process X t in R n, with t being the time parameter. Then, for f a twice continuously differentiable function with compact support, it is well known that t E[f(X t)] = E[L f(x t )], where E denotes the expectation with respect to the probability distribution of the diffusion process. At stationarity (i.e., t E[f(X t)] = 0), we therefore have E[L f(x t )] = 0, (2) and this equation leads to useful algebraic relations involving various expectations. Henceforward, we refer to (2) as the master equation. The key idea behind our work is to choose appropriate functions f in the master equation so that expectations of interest can be computed by solving systems of linear equations. Following Ohta and Kimura (1969a,b), we consider a diffusion process in a three-dimensional space parametrized by p, q and D (i.e., in the above notation, X t = (p, q, D) t ). The generator 5
6 corresponding to the two-locus model that we are considering in this paper is L = 1 2 p(1 p) 2 p q(1 q) 2 q [p(1 p)q(1 q) + D(1 2p)(1 2q) D2 ] 2 D 2 +D 2 p q + D(1 2p) 2 p D + D(1 2q) 2 q D + θ 4 (1 2p) p + θ 4 (1 2q) q D ( 1 + ρ 2 + θ ) D, (3) which differs from that of Ohta and Kimura (1969b) by a factor of 2; one unit of time corresponds to 2N e (rather than N e ) generations in our convention. 2.2 Reformulation of the problem: Summing over E[D 2 p m q n ] The main difficulty involved in computing the expectation of r 2 comes from the fact p(1 p)q(1 q) appears in the denominator. The strategy that we adopt in our work is to re-express (1) in terms of quantities whose expectations are easier to compute. First, note that ( 1 1 p(1 p)q(1 q) = p + 1 ) ( 1 1 p q + 1 ). 1 q Then, we can use the convergent series expansion 1/(1 z) = k=0 zk, where 0 z < 1, to obtain the following expressions for 0 < p < 1: 1 1 p = k=0 p k and 1 p = 1 1 (1 p) = (1 p) k. k=0 Similar results hold for 1/(1 q) and 1/q. Further, since E[r 2 ] is bounded, the Lebesgue convergence theorem implies [ E[r 2 D 2 ] ] = E p(1 p)q(1 q) = m=0 n=0 { E[D 2 p m q n ] + E[D 2 (1 p) m (1 q) n ] +E[D 2 (1 p) m q n ] + E[D 2 p m (1 q) n ] }. Finally, since E[D 2 p m q n ] = E[D 2 (1 p) m q n ] = E[D 2 p m (1 q) n ] = E[D 2 (1 p) m (1 q) n ] for the assumed model of mutation, [ E[r 2 D 2 ] ] = E p(1 p)q(1 q) = 4 m=0 n=0 E[D 2 p m q n ]. (4) We have thus translated the problem of computing E[r 2 ] into a problem of computing an infinite number of expectations of form E[D 2 p m q n ]. As we elaborate shortly, this change in perspective is 6
7 useful for two reasons: First, E[D 2 p m q n ] can be computed. Second, 4 m,n E[D2 p m q n ] converges fast as the sum m + n of exponents increases, thus allowing us to truncate the summation over m and n at some appropriate level. 2.3 Warm-up exercises Before we proceed to the computation of E[D 2 p m q n ], we here demonstrate how the diffusion generator technique works by computing some simpler expectations. These expectations are used in our algorithm for computing E[D 2 p m q n ]. The subsequent discussion pertains to the generator L given by (3). From using f = p n or f = q n in the master equation (2), it immediately follows that the expectations E[p n ] and E[q n ] satisfy E[p n ] = 2 + n 1 ) E[p n 1 ] and E[q n ] = θ + n 1 Since E[1] = 1, we can solve these recursions to obtain 2 + n 1 ) E[q n 1 ]. (5) θ + n 1 E[p n ] = E[q n ] = 2) [n], (6) [n] (θ) where (z) [k] denotes the rising factorial, defined as (z) [k] := z(z + 1) (z + k 1). (7) Using f = D in the master equation (2) implies E[D] = 0, while using f = Dp n, for n 1, yields the relation 2[2 + ρ + 2θ + n(3 + n + θ)]e[dp n ] = n(2 + 2n + θ)e[dp n 1 ]. A similar relation holds for E[Dq n ] and E[Dq n 1 ]. Hence, it follows from induction that E[Dp n ] = E[Dq n ] = 0 for all n 0. (8) If f = pq is used, we obtain θ E[pq] = E[D] + θ 4 (E[q] + E[p]). Since E[D] = 0 and E[p] = E[q] = 1/2, we conclude that E[pq] = 1 4. (9) Lastly, using f = p 2 q yields 2(2 + 3θ)E[p 2 q] = 8E[Dp] + θ E[p 2 ] + 2(2 + θ)e[pq], whereas using f = pq 2 yields 2(2 + 3θ)E[pq 2 ] = 8E[Dq] + θ E[q 2 ] + 2(2 + θ)e[pq]. Since E[Dp] = 0, E[p 2 ] = θ ) and E[pq] = 1 4, we conclude E[p 2 q] = E[pq 2 ] = 1 4 θ(θ+1), ). (10) θ + 1 7
8 n l = m Figure 1: Pairs (m, n) corresponding to m n, where m and n are exponents of p and q, respectively, in E[D 2 p m q n ]. A constant level l = m + n is indicated by a dashed line. Within each fixed level, open arrows indicate the order in which our algorithm is carried out. 2.4 Computing E[D 2 p m q n ] via solving systems of coupled linear equations All expectations considered in the previous section were quite straightforward to compute; by using a single appropriate function f in the master equation (2), we obtained a linear equation containing the expectation that we wished to compute and other expectations that had already been computed. Computing E[D 2 p m q n ] is more complicated, however, because the recursion relations now involve higher powers of D. We here present an algorithm for computing E[D 2 p m q n ] systematically. It involves solving systems of coupled linear equations in a particular order. First, note that by symmetry of the problem, p and q are exchangeable, i.e., E[D k p i q j ] = E[D k p j q i ], for all i, j, k 0. Without loss of generality, we may therefore assume that m n in E[D 2 p m q n ]. By a level we mean the sum l = m + n of the exponents of p and q in E[D 2 p m q n ]. This definition is depicted in Figure 1, where the pairs (m, n) corresponding to m n are indicated by closed circles. Our algorithm starts from level 0 and progresses upwards in level. As illustrated in Figure 1, within each fixed level l, we start from n = 0 and end at n = l/2 if l is even or at n = (l 1)/2 if l is odd. For each pair (m, n), where m n, we generate a system of n + 3 coupled linear equations by using f = D k p m+2 k q n+2 k in the master equation (2) for k = 0,..., n + 2. If computations are carried out in the particular order described above, the only unknown quantities in the system of coupled linear equations thus generated will be the following n + 3 expectations: E[p m+2 q n+2 ], E[Dp m+1 q n+1 ], E[D 2 p m q n ],..., E[D 2+n 1 p m n+1 q], E[D 2+n p m n ]. (11) 8
9 for l = 0,..., l max do if (l is even) then set n max = l/2 else set n max = (l 1)/2 end if for n = 0,..., n max do set m = l n. Then, set up a system of n + 3 coupled linear equations using f = D k p m+2 k q n+2 k in the master equation (2) for 0 k n + 2, and then solve for the n + 3 expectations E[D k p m+2 k q n+2 k ], 0 k n + 2 if (n m) then for 0 k n + 2 do set E[D k p n+2 k q m+2 k ] = E[D k p m+2 k q n+2 k ] end for end if end for end for Figure 2: Algorithm for computing E[D 2 p m q n ] up to some given truncation level l max. More precisely, if f = D k p i q j is used in the master equation (2), then in general we obtain [k 2 + i(i 1 + θ) + j(j 1 + θ) + k(1 + 4i + 4j + ρ + 2θ)] E[D k p i q j ] = 2ij E[D k+1 p i 1 q j 1 ] (2j2 + jθ + 4kj 2j) E[D k p i q j 1 ] (2i2 + iθ + 4ki 2i) E[D k p i 1 q j ] ( ) +k(k 1) 4 E[D k 1 p i+1 q j+1 ] + E[D k 1 p i q j ] 2 E[D k 1 p i q j+1 ] 2 E[D k 1 p i+1 q j ] ( ) +k(k 1) E[D k 2 p i+2 q j+2 ] + E[D k 2 p i+1 q j+1 ] E[D k 2 p i+2 q j+1 ] E[D k 2 p i+1 q j+2 ], where E[D k p i q j ], E[D k+1 p i 1 q j 1 ], E[D k 1 p i+1 q j+1 ], E[D k 2 p i+2 q j+2 ] are unknown quantities. We remark that the expectations shown in (6), (9), and (10) also appear in some equations and that they are treated as known quantities. Once we solve for the n + 3 expectations shown in (11), we move on to the next pair (m, n ) in order. A summary of the above algorithm is shown in Figure Level truncation and convergence In (4), m and n both range from 0 to. If we knew a closed-form formula for E[D 2 p m q n ], then it might be possible to obtain a closed-form formula for E[r 2 ] by summing over m and n explicitly. However, it seems quite difficult to obtain a closed-form formula for E[D 2 p m q n ], and therefore we have adopted a numerical approach. We have made two independent implementations of the algorithm described in Section 2.4: one 9
10 in C and the other in Mathematica. Both programs are available upon request. The Mathematica program can compute E[D 2 p m q n ] symbolically for given m and n, and can generate formulae in terms of θ and ρ. The C program is called ER2, and for given numerical values of θ, ρ, and l max, it computes the following level-truncated estimate of E[r 2 ]: E[r 2 ] lmax := l max l=0 a l, where a l := m, n 0, m + n = l 4 E[D 2 p m q n ]. (12) Since E[r 2 ] is bounded and the sequence {E[r 2 ] lmax } l max=0 of partial sums is a monotonically increasing sequence, {E[r 2 ] lmax } l max=0 is a convergent sequence. Although an analytic expression for the rate of convergence is difficult to obtain, we have empirically observed that the sequence converges quite fast. Shown in Figure 3a and Figure 3b are plots of E[r 2 ] lmax as a function of l max for ρ = 1 and ρ = 10, respectively. For given θ and ρ, ER2 took a few minutes on a laptop to compute E[r 2 ] lmax up to l max = 700. The rate of convergence of E[r 2 ] lmax seems to depend on θ and ρ; E[r 2 ] lmax converges faster for smaller ρ and larger θ. 2.6 Simplification: even from odd or odd from even The amount of computation involved in our algorithm can be reduced considerably using E[D l p m q n ] = ( 1) l E[D l p m (1 q) n ] = ( 1) l E[D l (1 p) m q n ], which is valid under the assumed model of mutation. This simple observation implies the following set of relations: [1 ( 1) n ] E[D 2k p m q n ] = [1 ( 1) m ] E[D 2k p m q n ] = n 1 ( ) n ( 1) j E[D 2k p m q j ], (13) j j=0 m 1 j=0 j=0 ( ) m ( 1) j E[D 2k p j q n ], (14) j n 1 ( ) n [1 + ( 1) n ] E[D 2k+1 p m q n ] = ( 1) j E[D 2k+1 p m q j ], (15) j [1 + ( 1) m ] E[D 2k+1 p m q n ] = m 1 j=0 ( ) m ( 1) j E[D 2k+1 p j q n ]. (16) j Relations (13) and (14) allow us to express E[D 2k p m q n ] for odd m or odd n purely in terms of E[D 2k p i q j ], where i m and j n are both even. In a similar vein, relations (15) and (16) allow 10
11 (a) 0.09 E[r 2 ]lmax θ = 1.0 θ = 0.8 θ = 0.6 θ = 0.4 θ = 0.2 θ = Truncation Level l max (b) E[r 2 ]lmax θ = 1.0 θ = 0.8 θ = 0.6 θ = 0.4 θ = 0.2 θ = Truncation Level l max Figure 3: Plots of level truncated estimate E[r 2 ] lmax, defined in (12), for various values of θ and ρ. (a) Plots for ρ = 1. (b) Plots for ρ = 10. For a given pair of θ and ρ, our software ER2 took a few minutes on a laptop to compute E[r 2 ] lmax up to l max = 700. As these plots show, the sequence {E[r 2 ] lmax } l max=0 of partial sums converges fast in general. The precise rate of convergence of depends on θ and ρ. 11
12 us to express E[D 2k+1 p m q n ] for even m or even n purely in terms of E[D 2k+1 p i q j ], where i m and j n are both odd. Relations (13) (16), together with the aforementioned observation that E[D l p m q n ] = E[D l p n q m ], significantly reduce the number of expectations that need to be computed explicitly. For example, it is straightforward to show that E[D 2k p 4 q 3 ] = E[D 2k p 3 q 4 ] = 1 4 E[D 2k p 3 q 3 ] = 1 16 ( ) 6 E[D 2k p 4 q 2 ] E[D 2k p 4 ], ( ) E[D 2k ] 12 E[D 2k p 2 ] + 36 E[D 2k p 2 q 2 ], E[D 2k+1 p 4 q 4 ] = 4 E[D 2k+1 p 3 q 3 ] 4 E[D 2k+1 p 3 q 1 ] + E[D 2k+1 pq], E[D 2k+1 p r ] = E[D 2k+1 q r ] = 0, for all r 0. Furthermore, E[r 2 ] lmax in (12) can be written purely in terms of E[D 2 p m q n ] in which both m and n are even, thus leading to a more efficient method of computing E[r 2 ] lmax. 3 Comparison of E[r 2 ] with σ 2 d and coalescent simulations In this section, we compare our computation of the expectation E[r 2 ] with averages of r 2 from coalescent simulations and also with the quantity σ 2 d = E[D 2 ] E[p(1 p)q(1 q)] = 10 + ρ + 4 θ ρ + ρ θρ + 32 θ + 8 θ 2, (17) obtained by Ohta and Kimura (1969b) under the same model as in the present paper. Previous simulation-based study, in the context of an infinitely-many-alleles model, has shown that σd 2 can be substantially larger than E[r 2 ] (Maruyama, 1982). Hudson (1985) has shown, however, that σd 2 is a reasonably good approximation of the sample estimated expectation of r 2 that is conditioned on minor allele frequencies being above 5%. The discussion below pertains to our assumed recurrent mutation model, taking all frequencies into account. Shown in Figure 4 is a plot of E[r 2 ] computed using our method, with l max = 700 as truncation level. A plot of σd 2 is shown in Figure 5a. As Figure 5b shows, E[r2 ] and σd 2 can be considerably different for certain parameter values. The figure shows that E[r 2 ] and σd 2 agree well for θ > 4. However, for small θ (say θ 1, which corresponds to a typical biologically interesting range), σd 2 can be larger than E[r 2 ] by a substantial amount (sometimes by tens of times), as in the aforementioned case of an infinitely-many-alleles model (Maruyama, 1982; Hudson, 1985). Numerical values of E[r 2 ] and σ 2 d are shown in Table 1a and Table 1b, respectively. For θ 1, E[r2 ] decreases as θ decreases, attaining negligibly small values for very small θ. In contrast, σd 2 can be very large even for very small θ. Note that σd 2 is a monotonically decreasing function of both θ and ρ. However, our computation indicates that, although E[r 2 ] is a monotonically decreasing function of ρ, it is not 12
13 E[r 2 ] ρ θ 8 10 Figure 4: A plot of E[r 2 ] computed using our method, truncated at level l max = 700. Parameters θ and ρ denote the population-scaled mutation and recombination rates, respectively. σ 2 d E[r 2 ] and σ 2 d ρ θ ρ θ 8 10 (a) (b) Figure 5: Comparison of σd 2 and our computation of E[r2 ]. (a) A plot of σd 2. (b) Superimposed plots of σd 2 and E[r2 ], the latter (in dotted lines) being the same as that shown in Figure 4. Note that E[r 2 ] and σd 2 exhibit very different behavior for certain parameter ranges. In particular, for θ < 1, σd 2 is larger than E[r2 ] by a substantial amount. a monotonically decreasing function of θ. For example, E[r 2 ] peaks at θ 1.5 for ρ = 0 and at θ 2.8 for ρ = 20. We also carried out coalescent simulations to estimate average r 2. We used Treevolve (available from Grassly et al. 1999) with a constant population size of 10 4, and performed at least 10, 000 simulations for each pair of ρ and θ considered. In every simulation, we generated 200 sequences each with exactly 2 sites. Mutation parameters were chosen to simulate a symmetric recurrent mutation model with two possible alleles per site. Table 1c shows a summary of average r 2 when no restriction is imposed on segregation. For θ 2, 13
14 they are much closer to our theoretical computation of E[r 2 ] (see Table 1a) than they are to σ 2 d (see Table 1b). As shown in Table 1d, however, when conditioned on segregation at both sites, sample estimated average r 2 increases substantially for small θ, resembling the behavior of σ 2 d. As Hudson (1985) noted, conditioning on minor allele frequencies being above 5% will make the sample estimated average of r 2 become even closer to σd 2. For large θ (say, θ 4), note that average r2 estimated from simulations tends to be slightly smaller than E[r 2 ] for ρ < 5, while being slightly larger than E[r 2 ] for large ρ (say, ρ > 10). 4 Asymptotic behavior: Large ρ limit Although a general closed-form formula for E[r 2 ] is difficult to obtain, it is possible to use our method to find relevant closed-form expressions in the limit ρ. It is easy to see from (17) that the asymptotic behavior of σd 2 is given by σ 2 d = 1 ρ + O(ρ 2 ) (Ohta and Kimura, 1969b). In this section, we use our method to show rigorously that the leading term of E[r 2 ] in the limit ρ also goes like 1/ρ, without any dependence on θ; i.e., E[r 2 ] = 1 ρ + O(ρ 2 ). 4.1 Asymptotic behavior of E[D 2 p m q n ] In our formulation (c.f., (4)), recall that computing E[r 2 ] amounts to computing E[D 2 p m q n ], for m, n 0. No closed-form formula for E[D 2 p m q n ] is known for arbitrary parameter values, and therefore we have constructed an algorithm that can be used to compute E[D 2 p m q n ] up to some chosen level l max = m + n. In the limit ρ, however, it turns out that we can obtain a closed-form formula for the leading term in the asymptotic expansion of E[D 2 p m q n ]. This analysis goes as follows: To find the asymptotic behavior E[D 2 p m q n ] = C(θ) ρ + O(ρ 2 ), 14
15 Table 1: Numerical comparison of E[r 2 ], σd 2, and average r2 from coalescent simulations. (a) Our computation of E[r 2 ] using l max = 700. (b) σd 2. (c) Average r2 from coalescent simulations, with no restriction on segregation. (d) Average r 2 from coalescent simulations, conditioned on segregation at both sites. In coalescent simulations, we used N e = 10 4 and generated 200 sequences each with exactly 2 sites. For each pair of ρ and θ, at least 10, 000 simulated data sets were used to compute the average r 2. For θ 4, almost all simulated data sets had segregation at both sites, thus explaining why (c) and (d) are the same for θ 4. (a) (b) (c) (d) θ ρ θ ρ θ ρ θ ρ
16 where C(θ) is some function of θ to be determined, let D 2 = ρd 2. Then, in the limit ρ, the master equation (2) can be written in terms of p, q, and D as [ p(1 p) 2 f E 2 p q(1 q) f q 2 + D(1 2p) 2 f p D + D(1 2q) 2 f q D + [ ρ pq(1 p)(1 q) + ρ D(1 2p)(1 2q) D 2] 2 f D θ (1 2p) f 4 p + θ (1 2q) f 4 q ρ f D 2 D ] = 0. (18) Substituting p m, q n, p m q n, Dp m q n and D 2 p m q n for f in (18) and letting ρ, we obtain the following recursion relations, respectively: E[p m ] = E[q n ] = 2 + m 1 ) E[p m 1 ], θ + m n 1 ) E[q n 1 ], θ + n 1 E[p m q n ] = 2 + m 1)E[pm 1 q n ] n 1)E[pm q n 1 ], m(θ + m 1) + n(θ + n 1) E[ Dp m q n ] = 0 E[ D 2 p m q n ] = E[p m+1 q n+1 ] E[p m+1 q n+2 ] E[p m+2 q n+1 ] + E[p m+2 q n+2 ]. Now, these recursions can be solved exactly; their solutions are given by E[p n ] = E[q n ] = 2) [n] (θ) [n], E[p m q n ] = E[p m ] E[q n ] = E[ D 2 p m q n ] = ( 1 θ 2 + n + 1 ) ( θ + n + 1 ) [n] [m] 2 2) (θ) [n] (θ) [m], 1 θ 2 + m + 1 ) θ + m + 1 E[p m+1 q n+1 ] = 1 4 ) [m+1] [n+1] 2 2) (θ + 1) [m+1] (θ + 1) [n+1], where (z) [k] denotes the rising factorial defined in (7). Since D 2 = D 2 /ρ, the last equation implies that, in the limit ρ, E[D 2 p m q n ] = 1 4 ) [m+1] [n+1] 2 2) 1 (θ + 1) [m+1] (θ + 1) [n+1] ρ + O(ρ 2 ). (19) 16
17 As a non-trivial check, consider E[D 2 p 4 q 2 ] as a rational function of ρ and θ; the numerator has degree 13 in ρ and degree 21 in θ, while the denominator has degree 14 in ρ and degree 22 in θ. In the limit ρ, one can show that the asymptotic behavior of this rational function is E[D 2 p 4 q 2 ] = θ2 (θ + 4)(θ + 6)(θ + 8) 1024 (θ + 5)(θ θ + 3) 2 1 ρ + O(ρ 2 ), which agrees with (19) when m = 4, n = 2. We used our Mathematica program to make similar checks for other values of m, n. 4.2 Asymptotic behavior of E[r 2 ] Since E[r 2 ] = 4 m,n E[D2 p m q n ], to study the behavior of E[r 2 ] in the limit ρ, we need to sum over m and n in (19). For M a non-negative integer, we prove the following formula in Appendix A: M m=0 2) [m+1] (θ + 1) [m+1] = ) [M+1]. (20) [M+1] (θ + 1) The right hand side of (20) can be written in terms of Γ-functions using the fact that (x + 1) [M+1] = Γ(x + M + 2)/Γ(x + 1). Taking the limit M and using Stirling s formula for the asymptotic expansion of Γ-functions, we obtain, for θ > 0, ( θ [m+1] 2) m=0 m=0 n=0 (θ + 1) [m+1] = 1. (21) Thus, in the limit ρ, equations (19) and (21) together imply E[r 2 ] = 4 E[D 2 p m q n ] = 1 ρ + O(ρ 2 ), in which the leading term is independent of θ. As mentioned before, this behavior agrees with the asymptotic limit of σd 2 = E[D2 ]/E[p(1 p)q(1 q)] found by Ohta and Kimura (1969b). 5 Discrete process In this section, we consider a description of the model in discrete time with a finite state space. This description was used by Hill and Robertson (1968) in their moment-generating matrix method, later extended by Ohta and Kimura (1969b) to include recurrent mutation. Although the discrete process is conceptually easy to grasp, computation in that framework is more complicated than in the diffusion approximation approach. Our goal in this section is to demonstrate that computing expectations in the discrete process can be facilitated by combinatorial techniques, and that such 17
18 computations may prove useful for studying the accuracy of the diffusion approximation approach. 5.1 Expected changes after one generation We use p and q to denote the marginal allele frequencies, and f ij to denote the frequency of the gametic type A i B j at the start of generation t. After mutation, the expected marginal allele frequencies are p u = (1 2u)p + u and q u = (1 2u)q + u, while the expected gametic frequencies f u ij are given by f00 u = f 00 + u(f 01 + f 10 ) 2uf 00, f01 u = f 01 + u(f 00 + f 11 ) 2uf 01, f10 u = f 10 + u(f 00 + f 11 ) 2uf 10, f11 u = f 11 + u(f 01 + f 10 ) 2uf 11. Note that, p u = f00 u + f 01 u and q u = f00 u + f 10 u. Expected gametic frequencies after recombination are given by f00 c = (1 c)f00 u + cp u q u, f01 c = (1 c)f01 u + cp u (1 q u ), f10 c = (1 c)f10 u + c(1 p u )q u, f11 c = (1 c)f11 u + c(1 p u )(1 q u ), and expected frequencies after sampling can be obtained from taking expectations with respect to the following multinomial probability density: P S (i, j, k 2N e ) = (2N e )! i!j!k!(2n e i j k)! (f c 00) i (f c 01) j (f c 10) k (f c 11) 2Ne i j k. In the remainder of this section, we use E S to denote the expectation with respect to the sampling probability P S (i, j, k 2N e ), and E to denote the expectation with respect to the joint distribution of marginal allele frequencies and linkage disequilibrium. 5.2 Exact computation of E[p l ] In the algorithm described in Section 2.4, the expectation E[p l ] appears in some linear equations, and our computation of the expectation E[r 2 ] depends on E[p l ]. The accuracy of E[p l ] therefore reflects 18
19 the accuracy of the diffusion process approximation. In what follows, we compute E[p l ] exactly in the discrete process setting and compare it with the answer from the diffusion approximation. We first describe our exact computation of E[p l ]. It is straightforward to show that the exponential generating function E S [e x(i+j) ] is given by 2N e E S [e x(i+j) ] = i=0 2N e i j=0 2N e i j k=0 e x(i+j) P S (i, j, k 2N e ) = [(f c 00 + f c 01)e x + f c 10 + f c 11)] 2Ne, which, using f c 00 + f c 01 + f c 10 + f c 11 = 1 and f c 00 + f c 01 = p u, can be simplified as E S [e x(i+j) ] = [(f c 00 + f c 01)(e x 1) + 1] 2Ne = [p u (e x 1) + 1] 2Ne. We denote by p the marginal allele frequency at locus 1 at the start of generation t + 1. Then, [ (I ) ] + J l [ E[(p ) l 1 l ] ] = E E S = E 2N e (2N e ) l x l E S[e x(i+j) ]. (22) x=0 We now describe how l E S [e x(i+j) ] x=0 can be computed exactly. First, define G(x) := p x l u (e x 1) + 1 (i.e., E S [e x(i+j) ] = [G(x)] 2Ne ) and for k j. Then, since H(j, k) := j x j [G(x)]k, (23) x=0 j x j [G(x)]k = k j 1 x j 1 { p u e x [G(x)] k 1} = k j 1 x j 1 { [G(x)] k (1 p u )[G(x)] k 1}, we see that H(j, k) satisfies the following recursion relation: H(j, k) = k[h(j 1, k) (1 p u )H(j 1, k 1)]. (24) The boundary condition for this recursion is H(1, k) = kp u, for all k 1, and, as shown in Appendix B, the solution is given by H(j, k) = j (k) [i] S(j, i)p i u, (25) i=1 where (z) [i] denotes the falling factorial, defined as (z) [i] := z(z 1) (z i + 1), (26) 19
20 and S(j, i) is the Stirling number of the second kind, defined as the number of partitions of {1, 2,..., j} into i non-empty subsets. At stationarity, E[(p ) l ] = E[p l ], and we can use (22), (23) and (25) to obtain E[p l ] = 1 (2N e ) l l (2N e ) [i] S(l, i) E[((1 2u)p + u) i ], (27) i=1 where we have substituted p u = (1 2u)p + u. After some rearrangement, this equation allows us to compute E[p l ] recursively. We remark that E[1] = 1 implies E[p] = 1/2 for all u and N e. 5.3 Approximation for small u and large N e We now consider a case in which u 1 and (1/N e ) 1, such that terms proportional to u i (1/N e ) j where i + j > 1 may be ignored. For small u, note that p k u uk(p k 1 2p k ) + p k. Expanding the right hand side of (27) up to first order in either u or 1/(2N e ), but not both, gives [( E[p l ] S(l, l) 1 1 ) ] l(l 1) E[p l ] + ul(e[p l 1 ] 2E[p l ]) + 1 S(l, l 1)E[p l 1 ], 2N e 2 2N e which, since S(l, l) = 1 and S(l, l 1) = l(l 1)/2, implies E[p l ] 2 + l 1 ) E[p l 1 ], (28) θ + l 1 which, in turn, implies E[p l ] 2) [l] / (θ) [l]. Similar results hold for E[q l ]. Note that (28) is precisely the result we have obtained in (5) using diffusion theory. This example well illustrates the power of diffusion approximation; relations like (5) can be obtained very easily in that approach, whereas they may require a lot of effort to derive in the discrete analogue. 5.4 Comparison of the exact and approximate solutions We now compare some results from the exact recursion (27) with that from the approximate recursion (28). Note that the former explicitly depends on the effective population size N e, whereas the latter does not. Table 2 shows that, for fixed θ, (28) becomes a better approximation to the exact recursion (27) as N e increases. Further, for small N e, (28) becomes a significantly better approximation as the scaled mutation rate θ decreases. For N e 10 4, (28) is in general a good approximation for all θ 1. 20
21 Table 2: Numerical values of E[p m ] obtained from the exact recursion (27) or the approximate recursion (28). Results obtained from (28), which has no explicit dependence on the effective population size N e, are equal to that from the diffusion process approximation. m θ Recursion Used Eq.(27), N e = Eq.(27), N e = Eq.(27), N e = Eq.(28) Eq.(27), N e = Eq.(27), N e = Eq.(27), N e = Eq.(28) Discussion Although we have considered a symmetric recurrent mutation model for simplicity, the technique developed in this paper can be generalized to other mutation models. In particular, for a case with non-symmetric mutation rates or with unequal mutation rates at different loci, it should be straightforward to devise an algorithm similar to that in Section 2.4. Further, it should be possible to generalize our method to include natural selection, as described in (Ohta and Kimura, 1969a). As we have shown in this paper, one can use our method to compute E[r 2 ] numerically for given θ and ρ. It would, of course, be desirable if we could obtain a closed-form formula for E[r 2 ]. We believe that that is not completely out of reach. We here conclude with a description of an alternative perspective that may prove useful for future work on deriving a closed-form formula for E[r 2 ]. If f = Dp m+1 q n+1 is used in the master equation (2), we obtain 4(1 + m)(1 + n) E[D 2 p m q n ] = 2[ρ + θ(4 + m + n) m 2 + n 2 + 5m + 5n] E[Dp m+1 q n+1 ] (1 + m)(4 + 2m + θ) E[Dp m q 1+n ] (1 + n)(4 + 2n + θ) E[Dp m+1 q n ]. (29) Further, using f = p k q l, one can show that E[Dp k 1 q l 1 ] = 1 { [k(k 1 + θ) + l(l 1 + θ)] E[p k q l ] 2kl ( k k 1 + θ ) ( E[p k 1 q l ] l l 1 + θ ) } E[p k q l 1 ]. (30) 2 2 Hence, if the joint expectation E[p i q j ] of powers of marginal frequencies are known, then E[D 2 p m q n ] can easily be computed using (29) and (30), and there would be no need to solve systems of coupled 21
22 equations as described in Section 2.4. At this point, however, we do not know how to obtain a general expression for E[p i q j ]. Using (6), we can obtain the following generating function for E[p k ] and E[q k ]: ( E[e αp ] = E[e αq ] = 2 θ 1 e α 1 2 α 2 θ α ) ( 1 2 I θ 1 Γ θ ), 2 where I ν (z) is the modified Bessel function of the first kind. generating function E[e αp+βq ] for E[p i q j ] has the form Moreover, we can show that the E[e αp+βq ] = E[e αp ] E[e βq ] h(α 2, β 2 ), (31) where h(α 2, β 2 ) is a symmetric function in α 2 and β 2. We believe that finding an explicit formula for h(α 2, β 2 ) is worthy of further research, since knowing the generating function E[e αp+βq ] may lead to a closed-form formula for E[r 2 ]. Acknowledgment We thank Charles H. Langley for helpful comments. This research is supported in part by grants CCF and IIS (YSS) from National Science Foundation. 22
23 References Ethier, S. N., Griffiths, R. C., On the two-locus sampling distribution. J. Math. Biol. 29, Golding, G. B., The sampling distribution of linkage disequilibrium. Genetics 108, Grassly, N. C., Harvey, P. H., Holmes, E. C., Population dynamics of HIV-1 inferred from gene sequences. Genetics 151, , (Software webpage: Hill, W. G., Robertson, A., Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, Hill, W. G., Weir, B. S., Maximum-likelihood estimation of gene location by linkage disequilibrium. Am. J. Hum. Genet. 54, Hudson, R. R., Sampling distribution of linkage disequilibrium under an infinite allele model without selection. Genetics 109, Hudson, R. R., 2001a. Linkage disequilibrium and recombination. In: Balding, D. J., Bishop, M., Canning, C. (Eds.), Handbook of Statistical Genetics. Wiley, pp Hudson, R. R., 2001b. Two-locus sampling distributions and their application. Genetics 159, International HapMap Consortium, A haplotype map of the human genome. Nature 437, Maruyama, T., Stochastic integrals and their application to population genetics. In: Kimura, M. (Ed.), Molecular Evolution, Protein Polymorphism and their Neutral Theory. Springer-Verlag, Berlin, pp McVean, G. A. T., A genealogical interpretation of linkage disequilibrium. Genetics 162, Ohta, T., Kimura, M., 1969a. Linkage disequilibrium due to random genetic drift. Genet. Res. Camb. 13, Ohta, T., Kimura, M., 1969b. Linkage disequilibrium at steady state determined by random genetic drift and recurrent mutation. Genetics 63, Pritchard, J. K., Przeworski, M., Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69,
24 Appendix A Proof of Equation (20) We prove the claim by induction. The lemma clearly holds for M = 0. Now, assume the case for M. Then, using the induction hypothesis, we have M+1 n=0 2) [n+1] (θ + 1) [n+1] = ) [M+1] 2) [M+2] (θ + 1) [M+1] + (θ + 1) [M+2] 2 + 1) [M+1] (θ + M + 2) 2) [M+2] = 1 (θ + 1) [M+2] = ) [M+1] (θ + M + 2 θ 2 ) (θ + 1) [M+2] 2 + 1) [M+2] = 1 (θ + 1) [M+2], which proves the claim. B Proof of Equation (25) Let X, Y and Z be finite sets of order x, y and z. Let Φ = {ϕ : X Y } be the set of all maps from X to Y and likewise for Ψ = {ψ : Y Z}, with order of Φ = y x and Ψ = z y. Now define R ϕ,ψ = {(a, b, c) X Y Z b = ϕ(a) and c = ψ(b)} for each pair ϕ Φ, ψ Ψ. Let R = {R ϕ,ψ ϕ Φ, ψ Ψ} and H z (x, y) := R. Note that R ϕ,ψ may be equal to R ϕ,ψ for ϕ ϕ and ψ ψ, and H z (x, y) counts the number of distinct sets R ϕ,ψ in R. Lemma B.1 H z (x, y) can be expressed as H z (x, y) = x (y) [i] S(x, i)z i, i=1 where S(x, i) denotes the Stirling number of the second kind. In particular, H z (1, y) = y z. Proof: We first partition R as R = R 1 R x where R i = {R ϕ,ψ ϕ(x) = i}. Then, there are z choices for mapping i preimages of ψ, (y) [i] choices for the i image of ϕ, and S(x, i) ways of partitioning X into kernels of ϕ. Hence, R i = (y) [i] S(x, i)z i. Summing over i thus gives the desired result. 24
25 Lemma B.2 H z (x, y) satisfies the recursion relation H z (x, y) = y[h z (x 1, y) + (z 1)H z (x 1, y 1)] with initial condition H z (1, y) = y z. Proof: Fix an element a X and c Z, and define X = X \ {a} and Z = Z \ {c}. Then, we can partition R as R = R 1 R 2 R 3, where R 1 = {R ϕ,ψ ϕ(a) ϕ( X)}, R 2 = {R ϕ,ψ ϕ(a) / ϕ( X), ψ(ϕ(a)) = c}, R 3 = {R ϕ,ψ ϕ(a) / ϕ( X), ψ(ϕ(a)) c}. Now, define R ϕ,ψ to be as before by replacing X by X. Then, to each of the H z (x 1, y) sets R ϕ,ψ, we can obtain y sets R ϕ,ψ by adjoining one element (a, b, ψ(b)) for b ϕ( X) and (a, b, c) for b / ϕ( X), and extending ϕ to ϕ so that ϕ(a) = b. Then, there exits a one-to-one correspondence between the set of all such sets obtained and R 1 R 2, and we thus see that R 1 R 2 = yh z (x 1, y). It is also easy to see that R 3 = y(z 1)H z (x 1, y 1). Note that even when z is not an integer, H z (x, y) still satisfies the recursion relation in Lemma B.2 as a purely algebraic relation. 25
Closed-form sampling formulas for the coalescent with recombination
 0 / 21 Closed-form sampling formulas for the coalescent with recombination Yun S. Song CS Division and Department of Statistics University of California, Berkeley September 7, 2009 Joint work with Paul
0 / 21 Closed-form sampling formulas for the coalescent with recombination Yun S. Song CS Division and Department of Statistics University of California, Berkeley September 7, 2009 Joint work with Paul
Stationary Distribution of the Linkage Disequilibrium Coefficient r 2
 Stationary Distribution of the Linkage Disequilibrium Coefficient r 2 Wei Zhang, Jing Liu, Rachel Fewster and Jesse Goodman Department of Statistics, The University of Auckland December 1, 2015 Overview
Stationary Distribution of the Linkage Disequilibrium Coefficient r 2 Wei Zhang, Jing Liu, Rachel Fewster and Jesse Goodman Department of Statistics, The University of Auckland December 1, 2015 Overview
p(d g A,g B )p(g B ), g B
 Supplementary Note Marginal effects for two-locus models Here we derive the marginal effect size of the three models given in Figure 1 of the main text. For each model we assume the two loci (A and B)
Supplementary Note Marginal effects for two-locus models Here we derive the marginal effect size of the three models given in Figure 1 of the main text. For each model we assume the two loci (A and B)
I of a gene sampled from a randomly mating popdation,
 Copyright 0 1987 by the Genetics Society of America Average Number of Nucleotide Differences in a From a Single Subpopulation: A Test for Population Subdivision Curtis Strobeck Department of Zoology, University
Copyright 0 1987 by the Genetics Society of America Average Number of Nucleotide Differences in a From a Single Subpopulation: A Test for Population Subdivision Curtis Strobeck Department of Zoology, University
Counting All Possible Ancestral Configurations of Sample Sequences in Population Genetics
 1 Counting All Possible Ancestral Configurations of Sample Sequences in Population Genetics Yun S. Song, Rune Lyngsø, and Jotun Hein Abstract Given a set D of input sequences, a genealogy for D can be
1 Counting All Possible Ancestral Configurations of Sample Sequences in Population Genetics Yun S. Song, Rune Lyngsø, and Jotun Hein Abstract Given a set D of input sequences, a genealogy for D can be
CLOSED-FORM ASYMPTOTIC SAMPLING DISTRIBUTIONS UNDER THE COALESCENT WITH RECOMBINATION FOR AN ARBITRARY NUMBER OF LOCI
 Adv. Appl. Prob. 44, 391 407 (01) Printed in Northern Ireland Applied Probability Trust 01 CLOSED-FORM ASYMPTOTIC SAMPLING DISTRIBUTIONS UNDER THE COALESCENT WITH RECOMBINATION FOR AN ARBITRARY NUMBER
Adv. Appl. Prob. 44, 391 407 (01) Printed in Northern Ireland Applied Probability Trust 01 CLOSED-FORM ASYMPTOTIC SAMPLING DISTRIBUTIONS UNDER THE COALESCENT WITH RECOMBINATION FOR AN ARBITRARY NUMBER
By Paul A. Jenkins and Yun S. Song, University of California, Berkeley July 26, 2010
 PADÉ APPROXIMANTS AND EXACT TWO-LOCUS SAMPLING DISTRIBUTIONS By Paul A. Jenkins and Yun S. Song, University of California, Berkeley July 26, 2010 For population genetics models with recombination, obtaining
PADÉ APPROXIMANTS AND EXACT TWO-LOCUS SAMPLING DISTRIBUTIONS By Paul A. Jenkins and Yun S. Song, University of California, Berkeley July 26, 2010 For population genetics models with recombination, obtaining
Problems for 3505 (2011)
 Problems for 505 (2011) 1. In the simplex of genotype distributions x + y + z = 1, for two alleles, the Hardy- Weinberg distributions x = p 2, y = 2pq, z = q 2 (p + q = 1) are characterized by y 2 = 4xz.
Problems for 505 (2011) 1. In the simplex of genotype distributions x + y + z = 1, for two alleles, the Hardy- Weinberg distributions x = p 2, y = 2pq, z = q 2 (p + q = 1) are characterized by y 2 = 4xz.
Infinite Continued Fractions
 Infinite Continued Fractions 8-5-200 The value of an infinite continued fraction [a 0 ; a, a 2, ] is lim c k, where c k is the k-th convergent k If [a 0 ; a, a 2, ] is an infinite continued fraction with
Infinite Continued Fractions 8-5-200 The value of an infinite continued fraction [a 0 ; a, a 2, ] is lim c k, where c k is the k-th convergent k If [a 0 ; a, a 2, ] is an infinite continued fraction with
AN ASYMPTOTIC SAMPLING FORMULA FOR THE COALESCENT WITH RECOMBINATION. By Paul A. Jenkins and Yun S. Song, University of California, Berkeley
 AN ASYMPTOTIC SAMPLING FORMULA FOR THE COALESCENT WITH RECOMBINATION By Paul A. Jenkins and Yun S. Song, University of California, Berkeley Ewens sampling formula (ESF) is a one-parameter family of probability
AN ASYMPTOTIC SAMPLING FORMULA FOR THE COALESCENT WITH RECOMBINATION By Paul A. Jenkins and Yun S. Song, University of California, Berkeley Ewens sampling formula (ESF) is a one-parameter family of probability
Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination)
 12/5/14 Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination) Linkage Disequilibrium Genealogical Interpretation of LD Association Mapping 1 Linkage and Recombination v linkage equilibrium ²
12/5/14 Chapter 6 Linkage Disequilibrium & Gene Mapping (Recombination) Linkage Disequilibrium Genealogical Interpretation of LD Association Mapping 1 Linkage and Recombination v linkage equilibrium ²
Haplotyping as Perfect Phylogeny: A direct approach
 Haplotyping as Perfect Phylogeny: A direct approach Vineet Bafna Dan Gusfield Giuseppe Lancia Shibu Yooseph February 7, 2003 Abstract A full Haplotype Map of the human genome will prove extremely valuable
Haplotyping as Perfect Phylogeny: A direct approach Vineet Bafna Dan Gusfield Giuseppe Lancia Shibu Yooseph February 7, 2003 Abstract A full Haplotype Map of the human genome will prove extremely valuable
Frequency Spectra and Inference in Population Genetics
 Frequency Spectra and Inference in Population Genetics Although coalescent models have come to play a central role in population genetics, there are some situations where genealogies may not lead to efficient
Frequency Spectra and Inference in Population Genetics Although coalescent models have come to play a central role in population genetics, there are some situations where genealogies may not lead to efficient
Genetic hitch-hiking in a subdivided population
 Genet. Res., Camb. (1998), 71, pp. 155 160. With 3 figures. Printed in the United Kingdom 1998 Cambridge University Press 155 Genetic hitch-hiking in a subdivided population MONTGOMERY SLATKIN* AND THOMAS
Genet. Res., Camb. (1998), 71, pp. 155 160. With 3 figures. Printed in the United Kingdom 1998 Cambridge University Press 155 Genetic hitch-hiking in a subdivided population MONTGOMERY SLATKIN* AND THOMAS
Population Genetics: a tutorial
 : a tutorial Institute for Science and Technology Austria ThRaSh 2014 provides the basic mathematical foundation of evolutionary theory allows a better understanding of experiments allows the development
: a tutorial Institute for Science and Technology Austria ThRaSh 2014 provides the basic mathematical foundation of evolutionary theory allows a better understanding of experiments allows the development
Population Genetics I. Bio
 Population Genetics I. Bio5488-2018 Don Conrad dconrad@genetics.wustl.edu Why study population genetics? Functional Inference Demographic inference: History of mankind is written in our DNA. We can learn
Population Genetics I. Bio5488-2018 Don Conrad dconrad@genetics.wustl.edu Why study population genetics? Functional Inference Demographic inference: History of mankind is written in our DNA. We can learn
A Principled Approach to Deriving Approximate Conditional Sampling. Distributions in Population Genetics Models with Recombination
 Genetics: Published Articles Ahead of Print, published on June 30, 2010 as 10.1534/genetics.110.117986 A Principled Approach to Deriving Approximate Conditional Sampling Distributions in Population Genetics
Genetics: Published Articles Ahead of Print, published on June 30, 2010 as 10.1534/genetics.110.117986 A Principled Approach to Deriving Approximate Conditional Sampling Distributions in Population Genetics
Computational Aspects of Aggregation in Biological Systems
 Computational Aspects of Aggregation in Biological Systems Vladik Kreinovich and Max Shpak University of Texas at El Paso, El Paso, TX 79968, USA vladik@utep.edu, mshpak@utep.edu Summary. Many biologically
Computational Aspects of Aggregation in Biological Systems Vladik Kreinovich and Max Shpak University of Texas at El Paso, El Paso, TX 79968, USA vladik@utep.edu, mshpak@utep.edu Summary. Many biologically
Mathematical Induction
 Chapter 6 Mathematical Induction 6.1 The Process of Mathematical Induction 6.1.1 Motivating Mathematical Induction Consider the sum of the first several odd integers. produce the following: 1 = 1 1 + 3
Chapter 6 Mathematical Induction 6.1 The Process of Mathematical Induction 6.1.1 Motivating Mathematical Induction Consider the sum of the first several odd integers. produce the following: 1 = 1 1 + 3
Practical Algebra. A Step-by-step Approach. Brought to you by Softmath, producers of Algebrator Software
 Practical Algebra A Step-by-step Approach Brought to you by Softmath, producers of Algebrator Software 2 Algebra e-book Table of Contents Chapter 1 Algebraic expressions 5 1 Collecting... like terms 5
Practical Algebra A Step-by-step Approach Brought to you by Softmath, producers of Algebrator Software 2 Algebra e-book Table of Contents Chapter 1 Algebraic expressions 5 1 Collecting... like terms 5
3.3 Accumulation Sequences
 3.3. ACCUMULATION SEQUENCES 25 3.3 Accumulation Sequences Overview. One of the most important mathematical ideas in calculus is that of an accumulation of change for physical quantities. As we have been
3.3. ACCUMULATION SEQUENCES 25 3.3 Accumulation Sequences Overview. One of the most important mathematical ideas in calculus is that of an accumulation of change for physical quantities. As we have been
Quantitative trait evolution with mutations of large effect
 Quantitative trait evolution with mutations of large effect May 1, 2014 Quantitative traits Traits that vary continuously in populations - Mass - Height - Bristle number (approx) Adaption - Low oxygen
Quantitative trait evolution with mutations of large effect May 1, 2014 Quantitative traits Traits that vary continuously in populations - Mass - Height - Bristle number (approx) Adaption - Low oxygen
Chapter Generating Functions
 Chapter 8.1.1-8.1.2. Generating Functions Prof. Tesler Math 184A Fall 2017 Prof. Tesler Ch. 8. Generating Functions Math 184A / Fall 2017 1 / 63 Ordinary Generating Functions (OGF) Let a n (n = 0, 1,...)
Chapter 8.1.1-8.1.2. Generating Functions Prof. Tesler Math 184A Fall 2017 Prof. Tesler Ch. 8. Generating Functions Math 184A / Fall 2017 1 / 63 Ordinary Generating Functions (OGF) Let a n (n = 0, 1,...)
Modelling Linkage Disequilibrium, And Identifying Recombination Hotspots Using SNP Data
 Modelling Linkage Disequilibrium, And Identifying Recombination Hotspots Using SNP Data Na Li and Matthew Stephens July 25, 2003 Department of Biostatistics, University of Washington, Seattle, WA 98195
Modelling Linkage Disequilibrium, And Identifying Recombination Hotspots Using SNP Data Na Li and Matthew Stephens July 25, 2003 Department of Biostatistics, University of Washington, Seattle, WA 98195
On prediction and density estimation Peter McCullagh University of Chicago December 2004
 On prediction and density estimation Peter McCullagh University of Chicago December 2004 Summary Having observed the initial segment of a random sequence, subsequent values may be predicted by calculating
On prediction and density estimation Peter McCullagh University of Chicago December 2004 Summary Having observed the initial segment of a random sequence, subsequent values may be predicted by calculating
MULTISECTION METHOD AND FURTHER FORMULAE FOR π. De-Yin Zheng
 Indian J. pure appl. Math., 39(: 37-55, April 008 c Printed in India. MULTISECTION METHOD AND FURTHER FORMULAE FOR π De-Yin Zheng Department of Mathematics, Hangzhou Normal University, Hangzhou 30036,
Indian J. pure appl. Math., 39(: 37-55, April 008 c Printed in India. MULTISECTION METHOD AND FURTHER FORMULAE FOR π De-Yin Zheng Department of Mathematics, Hangzhou Normal University, Hangzhou 30036,
are the q-versions of n, n! and . The falling factorial is (x) k = x(x 1)(x 2)... (x k + 1).
 Lecture A jacques@ucsd.edu Notation: N, R, Z, F, C naturals, reals, integers, a field, complex numbers. p(n), S n,, b(n), s n, partition numbers, Stirling of the second ind, Bell numbers, Stirling of the
Lecture A jacques@ucsd.edu Notation: N, R, Z, F, C naturals, reals, integers, a field, complex numbers. p(n), S n,, b(n), s n, partition numbers, Stirling of the second ind, Bell numbers, Stirling of the
Mathematical models in population genetics II
 Mathematical models in population genetics II Anand Bhaskar Evolutionary Biology and Theory of Computing Bootcamp January 1, 014 Quick recap Large discrete-time randomly mating Wright-Fisher population
Mathematical models in population genetics II Anand Bhaskar Evolutionary Biology and Theory of Computing Bootcamp January 1, 014 Quick recap Large discrete-time randomly mating Wright-Fisher population
Theorem 5.3. Let E/F, E = F (u), be a simple field extension. Then u is algebraic if and only if E/F is finite. In this case, [E : F ] = deg f u.
![Theorem 5.3. Let E/F, E = F (u), be a simple field extension. Then u is algebraic if and only if E/F is finite. In this case, [E : F ] = deg f u. Theorem 5.3. Let E/F, E = F (u), be a simple field extension. Then u is algebraic if and only if E/F is finite. In this case, [E : F ] = deg f u.](/thumbs/81/83685479.jpg) 5. Fields 5.1. Field extensions. Let F E be a subfield of the field E. We also describe this situation by saying that E is an extension field of F, and we write E/F to express this fact. If E/F is a field
5. Fields 5.1. Field extensions. Let F E be a subfield of the field E. We also describe this situation by saying that E is an extension field of F, and we write E/F to express this fact. If E/F is a field
ALGEBRA I CCR MATH STANDARDS
 RELATIONSHIPS BETWEEN QUANTITIES AND REASONING WITH EQUATIONS M.A1HS.1 M.A1HS.2 M.A1HS.3 M.A1HS.4 M.A1HS.5 M.A1HS.6 M.A1HS.7 M.A1HS.8 M.A1HS.9 M.A1HS.10 Reason quantitatively and use units to solve problems.
RELATIONSHIPS BETWEEN QUANTITIES AND REASONING WITH EQUATIONS M.A1HS.1 M.A1HS.2 M.A1HS.3 M.A1HS.4 M.A1HS.5 M.A1HS.6 M.A1HS.7 M.A1HS.8 M.A1HS.9 M.A1HS.10 Reason quantitatively and use units to solve problems.
Fall 2017 Test II review problems
 Fall 2017 Test II review problems Dr. Holmes October 18, 2017 This is a quite miscellaneous grab bag of relevant problems from old tests. Some are certainly repeated. 1. Give the complete addition and
Fall 2017 Test II review problems Dr. Holmes October 18, 2017 This is a quite miscellaneous grab bag of relevant problems from old tests. Some are certainly repeated. 1. Give the complete addition and
A fast estimate for the population recombination rate based on regression
 Genetics: Early Online, published on April 15, 213 as 1.1534/genetics.113.21 A fast estimate for the population recombination rate based on regression Kao Lin, Andreas Futschik, Haipeng Li CAS Key Laboratory
Genetics: Early Online, published on April 15, 213 as 1.1534/genetics.113.21 A fast estimate for the population recombination rate based on regression Kao Lin, Andreas Futschik, Haipeng Li CAS Key Laboratory
Population Genetics. with implications for Linkage Disequilibrium. Chiara Sabatti, Human Genetics 6357a Gonda
 1 Population Genetics with implications for Linkage Disequilibrium Chiara Sabatti, Human Genetics 6357a Gonda csabatti@mednet.ucla.edu 2 Hardy-Weinberg Hypotheses: infinite populations; no inbreeding;
1 Population Genetics with implications for Linkage Disequilibrium Chiara Sabatti, Human Genetics 6357a Gonda csabatti@mednet.ucla.edu 2 Hardy-Weinberg Hypotheses: infinite populations; no inbreeding;
The Sommerfeld Polynomial Method: Harmonic Oscillator Example
 Chemistry 460 Fall 2017 Dr. Jean M. Standard October 2, 2017 The Sommerfeld Polynomial Method: Harmonic Oscillator Example Scaling the Harmonic Oscillator Equation Recall the basic definitions of the harmonic
Chemistry 460 Fall 2017 Dr. Jean M. Standard October 2, 2017 The Sommerfeld Polynomial Method: Harmonic Oscillator Example Scaling the Harmonic Oscillator Equation Recall the basic definitions of the harmonic
CDM. Recurrences and Fibonacci. 20-fibonacci 2017/12/15 23:16. Terminology 4. Recurrence Equations 3. Solution and Asymptotics 6.
 CDM Recurrences and Fibonacci 1 Recurrence Equations Klaus Sutner Carnegie Mellon University Second Order 20-fibonacci 2017/12/15 23:16 The Fibonacci Monoid Recurrence Equations 3 Terminology 4 We can
CDM Recurrences and Fibonacci 1 Recurrence Equations Klaus Sutner Carnegie Mellon University Second Order 20-fibonacci 2017/12/15 23:16 The Fibonacci Monoid Recurrence Equations 3 Terminology 4 We can
Finite Fields: An introduction through exercises Jonathan Buss Spring 2014
 Finite Fields: An introduction through exercises Jonathan Buss Spring 2014 A typical course in abstract algebra starts with groups, and then moves on to rings, vector spaces, fields, etc. This sequence
Finite Fields: An introduction through exercises Jonathan Buss Spring 2014 A typical course in abstract algebra starts with groups, and then moves on to rings, vector spaces, fields, etc. This sequence
1.5.1 ESTIMATION OF HAPLOTYPE FREQUENCIES:
 .5. ESTIMATION OF HAPLOTYPE FREQUENCIES: Chapter - 8 For SNPs, alleles A j,b j at locus j there are 4 haplotypes: A A, A B, B A and B B frequencies q,q,q 3,q 4. Assume HWE at haplotype level. Only the
.5. ESTIMATION OF HAPLOTYPE FREQUENCIES: Chapter - 8 For SNPs, alleles A j,b j at locus j there are 4 haplotypes: A A, A B, B A and B B frequencies q,q,q 3,q 4. Assume HWE at haplotype level. Only the
Gene Pool Recombination in Genetic Algorithms
 Gene Pool Recombination in Genetic Algorithms Heinz Mühlenbein GMD 53754 St. Augustin Germany muehlenbein@gmd.de Hans-Michael Voigt T.U. Berlin 13355 Berlin Germany voigt@fb10.tu-berlin.de Abstract: A
Gene Pool Recombination in Genetic Algorithms Heinz Mühlenbein GMD 53754 St. Augustin Germany muehlenbein@gmd.de Hans-Michael Voigt T.U. Berlin 13355 Berlin Germany voigt@fb10.tu-berlin.de Abstract: A
DISCRETIZED CONFIGURATIONS AND PARTIAL PARTITIONS
 DISCRETIZED CONFIGURATIONS AND PARTIAL PARTITIONS AARON ABRAMS, DAVID GAY, AND VALERIE HOWER Abstract. We show that the discretized configuration space of k points in the n-simplex is homotopy equivalent
DISCRETIZED CONFIGURATIONS AND PARTIAL PARTITIONS AARON ABRAMS, DAVID GAY, AND VALERIE HOWER Abstract. We show that the discretized configuration space of k points in the n-simplex is homotopy equivalent
CDM. Recurrences and Fibonacci
 CDM Recurrences and Fibonacci Klaus Sutner Carnegie Mellon University 20-fibonacci 2017/12/15 23:16 1 Recurrence Equations Second Order The Fibonacci Monoid Recurrence Equations 3 We can define a sequence
CDM Recurrences and Fibonacci Klaus Sutner Carnegie Mellon University 20-fibonacci 2017/12/15 23:16 1 Recurrence Equations Second Order The Fibonacci Monoid Recurrence Equations 3 We can define a sequence
1 + lim. n n+1. f(x) = x + 1, x 1. and we check that f is increasing, instead. Using the quotient rule, we easily find that. 1 (x + 1) 1 x (x + 1) 2 =
 Chapter 5 Sequences and series 5. Sequences Definition 5. (Sequence). A sequence is a function which is defined on the set N of natural numbers. Since such a function is uniquely determined by its values
Chapter 5 Sequences and series 5. Sequences Definition 5. (Sequence). A sequence is a function which is defined on the set N of natural numbers. Since such a function is uniquely determined by its values
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin
 Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
k-protected VERTICES IN BINARY SEARCH TREES
 k-protected VERTICES IN BINARY SEARCH TREES MIKLÓS BÓNA Abstract. We show that for every k, the probability that a randomly selected vertex of a random binary search tree on n nodes is at distance k from
k-protected VERTICES IN BINARY SEARCH TREES MIKLÓS BÓNA Abstract. We show that for every k, the probability that a randomly selected vertex of a random binary search tree on n nodes is at distance k from
MATH 802: ENUMERATIVE COMBINATORICS ASSIGNMENT 2
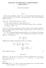 MATH 80: ENUMERATIVE COMBINATORICS ASSIGNMENT KANNAPPAN SAMPATH Facts Recall that, the Stirling number S(, n of the second ind is defined as the number of partitions of a [] into n non-empty blocs. We
MATH 80: ENUMERATIVE COMBINATORICS ASSIGNMENT KANNAPPAN SAMPATH Facts Recall that, the Stirling number S(, n of the second ind is defined as the number of partitions of a [] into n non-empty blocs. We
Lecture Notes Introduction to Ergodic Theory
 Lecture Notes Introduction to Ergodic Theory Tiago Pereira Department of Mathematics Imperial College London Our course consists of five introductory lectures on probabilistic aspects of dynamical systems,
Lecture Notes Introduction to Ergodic Theory Tiago Pereira Department of Mathematics Imperial College London Our course consists of five introductory lectures on probabilistic aspects of dynamical systems,
1 Springer. Nan M. Laird Christoph Lange. The Fundamentals of Modern Statistical Genetics
 1 Springer Nan M. Laird Christoph Lange The Fundamentals of Modern Statistical Genetics 1 Introduction to Statistical Genetics and Background in Molecular Genetics 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 Springer Nan M. Laird Christoph Lange The Fundamentals of Modern Statistical Genetics 1 Introduction to Statistical Genetics and Background in Molecular Genetics 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Supplemental Information Likelihood-based inference in isolation-by-distance models using the spatial distribution of low-frequency alleles
 Supplemental Information Likelihood-based inference in isolation-by-distance models using the spatial distribution of low-frequency alleles John Novembre and Montgomery Slatkin Supplementary Methods To
Supplemental Information Likelihood-based inference in isolation-by-distance models using the spatial distribution of low-frequency alleles John Novembre and Montgomery Slatkin Supplementary Methods To
This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and
 This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research education use, including for instruction at the authors institution
This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research education use, including for instruction at the authors institution
The Combinatorial Interpretation of Formulas in Coalescent Theory
 The Combinatorial Interpretation of Formulas in Coalescent Theory John L. Spouge National Center for Biotechnology Information NLM, NIH, DHHS spouge@ncbi.nlm.nih.gov Bldg. A, Rm. N 0 NCBI, NLM, NIH Bethesda
The Combinatorial Interpretation of Formulas in Coalescent Theory John L. Spouge National Center for Biotechnology Information NLM, NIH, DHHS spouge@ncbi.nlm.nih.gov Bldg. A, Rm. N 0 NCBI, NLM, NIH Bethesda
Problem Solving in Math (Math 43900) Fall 2013
 Problem Solving in Math (Math 43900) Fall 203 Week six (October ) problems recurrences Instructor: David Galvin Definition of a recurrence relation We met recurrences in the induction hand-out. Sometimes
Problem Solving in Math (Math 43900) Fall 203 Week six (October ) problems recurrences Instructor: David Galvin Definition of a recurrence relation We met recurrences in the induction hand-out. Sometimes
A comparison of two popular statistical methods for estimating the time to most recent common ancestor (TMRCA) from a sample of DNA sequences
 Indian Academy of Sciences A comparison of two popular statistical methods for estimating the time to most recent common ancestor (TMRCA) from a sample of DNA sequences ANALABHA BASU and PARTHA P. MAJUMDER*
Indian Academy of Sciences A comparison of two popular statistical methods for estimating the time to most recent common ancestor (TMRCA) from a sample of DNA sequences ANALABHA BASU and PARTHA P. MAJUMDER*
Contrasts for a within-species comparative method
 Contrasts for a within-species comparative method Joseph Felsenstein, Department of Genetics, University of Washington, Box 357360, Seattle, Washington 98195-7360, USA email address: joe@genetics.washington.edu
Contrasts for a within-species comparative method Joseph Felsenstein, Department of Genetics, University of Washington, Box 357360, Seattle, Washington 98195-7360, USA email address: joe@genetics.washington.edu
TOPOLOGICAL COMPLEXITY OF 2-TORSION LENS SPACES AND ku-(co)homology
 TOPOLOGICAL COMPLEXITY OF 2-TORSION LENS SPACES AND ku-(co)homology DONALD M. DAVIS Abstract. We use ku-cohomology to determine lower bounds for the topological complexity of mod-2 e lens spaces. In the
TOPOLOGICAL COMPLEXITY OF 2-TORSION LENS SPACES AND ku-(co)homology DONALD M. DAVIS Abstract. We use ku-cohomology to determine lower bounds for the topological complexity of mod-2 e lens spaces. In the
arxiv: v2 [q-bio.pe] 26 May 2011
![arxiv: v2 [q-bio.pe] 26 May 2011 arxiv: v2 [q-bio.pe] 26 May 2011](/thumbs/94/119031012.jpg) The Structure of Genealogies in the Presence of Purifying Selection: A Fitness-Class Coalescent arxiv:1010.2479v2 [q-bio.pe] 26 May 2011 Aleksandra M. Walczak 1,, Lauren E. Nicolaisen 2,, Joshua B. Plotkin
The Structure of Genealogies in the Presence of Purifying Selection: A Fitness-Class Coalescent arxiv:1010.2479v2 [q-bio.pe] 26 May 2011 Aleksandra M. Walczak 1,, Lauren E. Nicolaisen 2,, Joshua B. Plotkin
Mathematical Structures Combinations and Permutations
 Definitions: Suppose S is a (finite) set and n, k 0 are integers The set C(S, k) of k - combinations consists of all subsets of S that have exactly k elements The set P (S, k) of k - permutations consists
Definitions: Suppose S is a (finite) set and n, k 0 are integers The set C(S, k) of k - combinations consists of all subsets of S that have exactly k elements The set P (S, k) of k - permutations consists
URN MODELS: the Ewens Sampling Lemma
 Department of Computer Science Brown University, Providence sorin@cs.brown.edu October 3, 2014 1 2 3 4 Mutation Mutation: typical values for parameters Equilibrium Probability of fixation 5 6 Ewens Sampling
Department of Computer Science Brown University, Providence sorin@cs.brown.edu October 3, 2014 1 2 3 4 Mutation Mutation: typical values for parameters Equilibrium Probability of fixation 5 6 Ewens Sampling
Introduction. Chapter 1
 Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
Chapter 1 Introduction In this book we will be concerned with supervised learning, which is the problem of learning input-output mappings from empirical data (the training dataset). Depending on the characteristics
EGYPTIAN FRACTIONS WITH EACH DENOMINATOR HAVING THREE DISTINCT PRIME DIVISORS
 #A5 INTEGERS 5 (205) EGYPTIAN FRACTIONS WITH EACH DENOMINATOR HAVING THREE DISTINCT PRIME DIVISORS Steve Butler Department of Mathematics, Iowa State University, Ames, Iowa butler@iastate.edu Paul Erdős
#A5 INTEGERS 5 (205) EGYPTIAN FRACTIONS WITH EACH DENOMINATOR HAVING THREE DISTINCT PRIME DIVISORS Steve Butler Department of Mathematics, Iowa State University, Ames, Iowa butler@iastate.edu Paul Erdős
Spring 2012 Math 541B Exam 1
 Spring 2012 Math 541B Exam 1 1. A sample of size n is drawn without replacement from an urn containing N balls, m of which are red and N m are black; the balls are otherwise indistinguishable. Let X denote
Spring 2012 Math 541B Exam 1 1. A sample of size n is drawn without replacement from an urn containing N balls, m of which are red and N m are black; the balls are otherwise indistinguishable. Let X denote
Building Infinite Processes from Finite-Dimensional Distributions
 Chapter 2 Building Infinite Processes from Finite-Dimensional Distributions Section 2.1 introduces the finite-dimensional distributions of a stochastic process, and shows how they determine its infinite-dimensional
Chapter 2 Building Infinite Processes from Finite-Dimensional Distributions Section 2.1 introduces the finite-dimensional distributions of a stochastic process, and shows how they determine its infinite-dimensional
Induction and Recursion
 . All rights reserved. Authorized only for instructor use in the classroom. No reproduction or further distribution permitted without the prior written consent of McGraw-Hill Education. Induction and Recursion
. All rights reserved. Authorized only for instructor use in the classroom. No reproduction or further distribution permitted without the prior written consent of McGraw-Hill Education. Induction and Recursion
The Multivariate Gaussian Distribution [DRAFT]
![The Multivariate Gaussian Distribution [DRAFT] The Multivariate Gaussian Distribution [DRAFT]](/thumbs/90/102238180.jpg) The Multivariate Gaussian Distribution DRAFT David S. Rosenberg Abstract This is a collection of a few key and standard results about multivariate Gaussian distributions. I have not included many proofs,
The Multivariate Gaussian Distribution DRAFT David S. Rosenberg Abstract This is a collection of a few key and standard results about multivariate Gaussian distributions. I have not included many proofs,
Surfing genes. On the fate of neutral mutations in a spreading population
 Surfing genes On the fate of neutral mutations in a spreading population Oskar Hallatschek David Nelson Harvard University ohallats@physics.harvard.edu Genetic impact of range expansions Population expansions
Surfing genes On the fate of neutral mutations in a spreading population Oskar Hallatschek David Nelson Harvard University ohallats@physics.harvard.edu Genetic impact of range expansions Population expansions
Classical transcendental curves
 Classical transcendental curves Reinhard Schultz May, 2008 In his writings on coordinate geometry, Descartes emphasized that he was only willing to work with curves that could be defined by algebraic equations.
Classical transcendental curves Reinhard Schultz May, 2008 In his writings on coordinate geometry, Descartes emphasized that he was only willing to work with curves that could be defined by algebraic equations.
1 Basic Combinatorics
 1 Basic Combinatorics 1.1 Sets and sequences Sets. A set is an unordered collection of distinct objects. The objects are called elements of the set. We use braces to denote a set, for example, the set
1 Basic Combinatorics 1.1 Sets and sequences Sets. A set is an unordered collection of distinct objects. The objects are called elements of the set. We use braces to denote a set, for example, the set
Graphs with few total dominating sets
 Graphs with few total dominating sets Marcin Krzywkowski marcin.krzywkowski@gmail.com Stephan Wagner swagner@sun.ac.za Abstract We give a lower bound for the number of total dominating sets of a graph
Graphs with few total dominating sets Marcin Krzywkowski marcin.krzywkowski@gmail.com Stephan Wagner swagner@sun.ac.za Abstract We give a lower bound for the number of total dominating sets of a graph
CHAPTER 8: EXPLORING R
 CHAPTER 8: EXPLORING R LECTURE NOTES FOR MATH 378 (CSUSM, SPRING 2009). WAYNE AITKEN In the previous chapter we discussed the need for a complete ordered field. The field Q is not complete, so we constructed
CHAPTER 8: EXPLORING R LECTURE NOTES FOR MATH 378 (CSUSM, SPRING 2009). WAYNE AITKEN In the previous chapter we discussed the need for a complete ordered field. The field Q is not complete, so we constructed
Asymptotics of Integrals of. Hermite Polynomials
 Applied Mathematical Sciences, Vol. 4, 010, no. 61, 04-056 Asymptotics of Integrals of Hermite Polynomials R. B. Paris Division of Complex Systems University of Abertay Dundee Dundee DD1 1HG, UK R.Paris@abertay.ac.uk
Applied Mathematical Sciences, Vol. 4, 010, no. 61, 04-056 Asymptotics of Integrals of Hermite Polynomials R. B. Paris Division of Complex Systems University of Abertay Dundee Dundee DD1 1HG, UK R.Paris@abertay.ac.uk
SWEEPFINDER2: Increased sensitivity, robustness, and flexibility
 SWEEPFINDER2: Increased sensitivity, robustness, and flexibility Michael DeGiorgio 1,*, Christian D. Huber 2, Melissa J. Hubisz 3, Ines Hellmann 4, and Rasmus Nielsen 5 1 Department of Biology, Pennsylvania
SWEEPFINDER2: Increased sensitivity, robustness, and flexibility Michael DeGiorgio 1,*, Christian D. Huber 2, Melissa J. Hubisz 3, Ines Hellmann 4, and Rasmus Nielsen 5 1 Department of Biology, Pennsylvania
L(x; 0) = f(x) This family is equivalent to that obtained by the heat or diffusion equation: L t = 1 2 L
 Figure 1: Fine To Coarse Scales. The parameter t is often associated with the generation of a scale-space family, where t = 0 denotes the original image and t > 0 increasing (coarser) scales. Scale-Space
Figure 1: Fine To Coarse Scales. The parameter t is often associated with the generation of a scale-space family, where t = 0 denotes the original image and t > 0 increasing (coarser) scales. Scale-Space
Mathematics. Number and Quantity The Real Number System
 Number and Quantity The Real Number System Extend the properties of exponents to rational exponents. 1. Explain how the definition of the meaning of rational exponents follows from extending the properties
Number and Quantity The Real Number System Extend the properties of exponents to rational exponents. 1. Explain how the definition of the meaning of rational exponents follows from extending the properties
The Banach-Tarski paradox
 The Banach-Tarski paradox 1 Non-measurable sets In these notes I want to present a proof of the Banach-Tarski paradox, a consequence of the axiom of choice that shows us that a naive understanding of the
The Banach-Tarski paradox 1 Non-measurable sets In these notes I want to present a proof of the Banach-Tarski paradox, a consequence of the axiom of choice that shows us that a naive understanding of the
EVOLUTIONARY DYNAMICS AND THE EVOLUTION OF MULTIPLAYER COOPERATION IN A SUBDIVIDED POPULATION
 Friday, July 27th, 11:00 EVOLUTIONARY DYNAMICS AND THE EVOLUTION OF MULTIPLAYER COOPERATION IN A SUBDIVIDED POPULATION Karan Pattni karanp@liverpool.ac.uk University of Liverpool Joint work with Prof.
Friday, July 27th, 11:00 EVOLUTIONARY DYNAMICS AND THE EVOLUTION OF MULTIPLAYER COOPERATION IN A SUBDIVIDED POPULATION Karan Pattni karanp@liverpool.ac.uk University of Liverpool Joint work with Prof.
Introduction to Divide and Conquer
 Introduction to Divide and Conquer Sorting with O(n log n) comparisons and integer multiplication faster than O(n 2 ) Periklis A. Papakonstantinou York University Consider a problem that admits a straightforward
Introduction to Divide and Conquer Sorting with O(n log n) comparisons and integer multiplication faster than O(n 2 ) Periklis A. Papakonstantinou York University Consider a problem that admits a straightforward
CS281B / Stat 241B : Statistical Learning Theory Lecture: #22 on 19 Apr Dirichlet Process I
 X i Ν CS281B / Stat 241B : Statistical Learning Theory Lecture: #22 on 19 Apr 2004 Dirichlet Process I Lecturer: Prof. Michael Jordan Scribe: Daniel Schonberg dschonbe@eecs.berkeley.edu 22.1 Dirichlet
X i Ν CS281B / Stat 241B : Statistical Learning Theory Lecture: #22 on 19 Apr 2004 Dirichlet Process I Lecturer: Prof. Michael Jordan Scribe: Daniel Schonberg dschonbe@eecs.berkeley.edu 22.1 Dirichlet
arxiv: v1 [math.nt] 20 Mar 2017
![arxiv: v1 [math.nt] 20 Mar 2017 arxiv: v1 [math.nt] 20 Mar 2017](/thumbs/90/101804034.jpg) On certain ratios regarding integer numbers which are both triangulars and squares arxiv:1703.06701v1 [math.nt] 0 Mar 017 Fabio Roman Abstract We investigate integer numbers which possess at the same time
On certain ratios regarding integer numbers which are both triangulars and squares arxiv:1703.06701v1 [math.nt] 0 Mar 017 Fabio Roman Abstract We investigate integer numbers which possess at the same time
Estimating effective population size from samples of sequences: inefficiency of pairwise and segregating sites as compared to phylogenetic estimates
 Estimating effective population size from samples of sequences: inefficiency of pairwise and segregating sites as compared to phylogenetic estimates JOSEPH FELSENSTEIN Department of Genetics SK-50, University
Estimating effective population size from samples of sequences: inefficiency of pairwise and segregating sites as compared to phylogenetic estimates JOSEPH FELSENSTEIN Department of Genetics SK-50, University
ALGEBRA I. 2. Rewrite expressions involving radicals and rational exponents using the properties of exponents. (N-RN2)
 ALGEBRA I The Algebra I course builds on foundational mathematical content learned by students in Grades K-8 by expanding mathematics understanding to provide students with a strong mathematics education.
ALGEBRA I The Algebra I course builds on foundational mathematical content learned by students in Grades K-8 by expanding mathematics understanding to provide students with a strong mathematics education.
Elements with Square Roots in Finite Groups
 Elements with Square Roots in Finite Groups M. S. Lucido, M. R. Pournaki * Abstract In this paper, we study the probability that a randomly chosen element in a finite group has a square root, in particular
Elements with Square Roots in Finite Groups M. S. Lucido, M. R. Pournaki * Abstract In this paper, we study the probability that a randomly chosen element in a finite group has a square root, in particular
Mathematics High School Algebra I
 Mathematics High School Algebra I All West Virginia teachers are responsible for classroom instruction that integrates content standards and mathematical habits of mind. Students in this course will focus
Mathematics High School Algebra I All West Virginia teachers are responsible for classroom instruction that integrates content standards and mathematical habits of mind. Students in this course will focus
Hardy martingales and Jensen s Inequality
 Hardy martingales and Jensen s Inequality Nakhlé H. Asmar and Stephen J. Montgomery Smith Department of Mathematics University of Missouri Columbia Columbia, Missouri 65211 U. S. A. Abstract Hardy martingales
Hardy martingales and Jensen s Inequality Nakhlé H. Asmar and Stephen J. Montgomery Smith Department of Mathematics University of Missouri Columbia Columbia, Missouri 65211 U. S. A. Abstract Hardy martingales
Generating Functions
 Semester 1, 2004 Generating functions Another means of organising enumeration. Two examples we have seen already. Example 1. Binomial coefficients. Let X = {1, 2,..., n} c k = # k-element subsets of X
Semester 1, 2004 Generating functions Another means of organising enumeration. Two examples we have seen already. Example 1. Binomial coefficients. Let X = {1, 2,..., n} c k = # k-element subsets of X
Lecture 24: Multivariate Response: Changes in G. Bruce Walsh lecture notes Synbreed course version 10 July 2013
 Lecture 24: Multivariate Response: Changes in G Bruce Walsh lecture notes Synbreed course version 10 July 2013 1 Overview Changes in G from disequilibrium (generalized Bulmer Equation) Fragility of covariances
Lecture 24: Multivariate Response: Changes in G Bruce Walsh lecture notes Synbreed course version 10 July 2013 1 Overview Changes in G from disequilibrium (generalized Bulmer Equation) Fragility of covariances
ON COMPOUND POISSON POPULATION MODELS
 ON COMPOUND POISSON POPULATION MODELS Martin Möhle, University of Tübingen (joint work with Thierry Huillet, Université de Cergy-Pontoise) Workshop on Probability, Population Genetics and Evolution Centre
ON COMPOUND POISSON POPULATION MODELS Martin Möhle, University of Tübingen (joint work with Thierry Huillet, Université de Cergy-Pontoise) Workshop on Probability, Population Genetics and Evolution Centre
Quadratic reciprocity (after Weil) 1. Standard set-up and Poisson summation
 (December 19, 010 Quadratic reciprocity (after Weil Paul Garrett garrett@math.umn.edu http://www.math.umn.edu/ garrett/ I show that over global fields k (characteristic not the quadratic norm residue symbol
(December 19, 010 Quadratic reciprocity (after Weil Paul Garrett garrett@math.umn.edu http://www.math.umn.edu/ garrett/ I show that over global fields k (characteristic not the quadratic norm residue symbol
arxiv: v1 [math.co] 21 Sep 2015
![arxiv: v1 [math.co] 21 Sep 2015 arxiv: v1 [math.co] 21 Sep 2015](/thumbs/84/89744698.jpg) Chocolate Numbers arxiv:1509.06093v1 [math.co] 21 Sep 2015 Caleb Ji, Tanya Khovanova, Robin Park, Angela Song September 22, 2015 Abstract In this paper, we consider a game played on a rectangular m n gridded
Chocolate Numbers arxiv:1509.06093v1 [math.co] 21 Sep 2015 Caleb Ji, Tanya Khovanova, Robin Park, Angela Song September 22, 2015 Abstract In this paper, we consider a game played on a rectangular m n gridded
Random Times and Their Properties
 Chapter 6 Random Times and Their Properties Section 6.1 recalls the definition of a filtration (a growing collection of σ-fields) and of stopping times (basically, measurable random times). Section 6.2
Chapter 6 Random Times and Their Properties Section 6.1 recalls the definition of a filtration (a growing collection of σ-fields) and of stopping times (basically, measurable random times). Section 6.2
FINITE CONNECTED H-SPACES ARE CONTRACTIBLE
 FINITE CONNECTED H-SPACES ARE CONTRACTIBLE ISAAC FRIEND Abstract. The non-hausdorff suspension of the one-sphere S 1 of complex numbers fails to model the group s continuous multiplication. Moreover, finite
FINITE CONNECTED H-SPACES ARE CONTRACTIBLE ISAAC FRIEND Abstract. The non-hausdorff suspension of the one-sphere S 1 of complex numbers fails to model the group s continuous multiplication. Moreover, finite
Defining the Integral
 Defining the Integral In these notes we provide a careful definition of the Lebesgue integral and we prove each of the three main convergence theorems. For the duration of these notes, let (, M, µ) be
Defining the Integral In these notes we provide a careful definition of the Lebesgue integral and we prove each of the three main convergence theorems. For the duration of these notes, let (, M, µ) be
Walker Ray Econ 204 Problem Set 3 Suggested Solutions August 6, 2015
 Problem 1. Take any mapping f from a metric space X into a metric space Y. Prove that f is continuous if and only if f(a) f(a). (Hint: use the closed set characterization of continuity). I make use of
Problem 1. Take any mapping f from a metric space X into a metric space Y. Prove that f is continuous if and only if f(a) f(a). (Hint: use the closed set characterization of continuity). I make use of
The Structure of Genealogies in the Presence of Purifying Selection: a "Fitness-Class Coalescent"
 The Structure of Genealogies in the Presence of Purifying Selection: a "Fitness-Class Coalescent" The Harvard community has made this article openly available. Please share how this access benefits you.
The Structure of Genealogies in the Presence of Purifying Selection: a "Fitness-Class Coalescent" The Harvard community has made this article openly available. Please share how this access benefits you.
Derangements with an additional restriction (and zigzags)
 Derangements with an additional restriction (and zigzags) István Mező Nanjing University of Information Science and Technology 2017. 05. 27. This talk is about a class of generalized derangement numbers,
Derangements with an additional restriction (and zigzags) István Mező Nanjing University of Information Science and Technology 2017. 05. 27. This talk is about a class of generalized derangement numbers,
Lebesgue Measure on R n
 CHAPTER 2 Lebesgue Measure on R n Our goal is to construct a notion of the volume, or Lebesgue measure, of rather general subsets of R n that reduces to the usual volume of elementary geometrical sets
CHAPTER 2 Lebesgue Measure on R n Our goal is to construct a notion of the volume, or Lebesgue measure, of rather general subsets of R n that reduces to the usual volume of elementary geometrical sets
On rational approximation of algebraic functions. Julius Borcea. Rikard Bøgvad & Boris Shapiro
 On rational approximation of algebraic functions http://arxiv.org/abs/math.ca/0409353 Julius Borcea joint work with Rikard Bøgvad & Boris Shapiro 1. Padé approximation: short overview 2. A scheme of rational
On rational approximation of algebraic functions http://arxiv.org/abs/math.ca/0409353 Julius Borcea joint work with Rikard Bøgvad & Boris Shapiro 1. Padé approximation: short overview 2. A scheme of rational
Course 311: Michaelmas Term 2005 Part III: Topics in Commutative Algebra
 Course 311: Michaelmas Term 2005 Part III: Topics in Commutative Algebra D. R. Wilkins Contents 3 Topics in Commutative Algebra 2 3.1 Rings and Fields......................... 2 3.2 Ideals...............................
Course 311: Michaelmas Term 2005 Part III: Topics in Commutative Algebra D. R. Wilkins Contents 3 Topics in Commutative Algebra 2 3.1 Rings and Fields......................... 2 3.2 Ideals...............................
Chapter Summary. Mathematical Induction Strong Induction Well-Ordering Recursive Definitions Structural Induction Recursive Algorithms
 1 Chapter Summary Mathematical Induction Strong Induction Well-Ordering Recursive Definitions Structural Induction Recursive Algorithms 2 Section 5.1 3 Section Summary Mathematical Induction Examples of
1 Chapter Summary Mathematical Induction Strong Induction Well-Ordering Recursive Definitions Structural Induction Recursive Algorithms 2 Section 5.1 3 Section Summary Mathematical Induction Examples of
Abstract. 2. We construct several transcendental numbers.
 Abstract. We prove Liouville s Theorem for the order of approximation by rationals of real algebraic numbers. 2. We construct several transcendental numbers. 3. We define Poissonian Behaviour, and study
Abstract. We prove Liouville s Theorem for the order of approximation by rationals of real algebraic numbers. 2. We construct several transcendental numbers. 3. We define Poissonian Behaviour, and study
A Combinatorial Interpretation of the Numbers 6 (2n)! /n! (n + 2)!
 1 2 3 47 6 23 11 Journal of Integer Sequences, Vol. 8 (2005), Article 05.2.3 A Combinatorial Interpretation of the Numbers 6 (2n)! /n! (n + 2)! Ira M. Gessel 1 and Guoce Xin Department of Mathematics Brandeis
1 2 3 47 6 23 11 Journal of Integer Sequences, Vol. 8 (2005), Article 05.2.3 A Combinatorial Interpretation of the Numbers 6 (2n)! /n! (n + 2)! Ira M. Gessel 1 and Guoce Xin Department of Mathematics Brandeis
Statistics 581 Revision of Section 4.4: Consistency of Maximum Likelihood Estimates Wellner; 11/30/2001
 Statistics 581 Revision of Section 4.4: Consistency of Maximum Likelihood Estimates Wellner; 11/30/2001 Some Uniform Strong Laws of Large Numbers Suppose that: A. X, X 1,...,X n are i.i.d. P on the measurable
Statistics 581 Revision of Section 4.4: Consistency of Maximum Likelihood Estimates Wellner; 11/30/2001 Some Uniform Strong Laws of Large Numbers Suppose that: A. X, X 1,...,X n are i.i.d. P on the measurable
Outline of lectures 3-6
 GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 009 Population genetics Outline of lectures 3-6 1. We want to know what theory says about the reproduction of genotypes in a population. This results
GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 009 Population genetics Outline of lectures 3-6 1. We want to know what theory says about the reproduction of genotypes in a population. This results
