Online Appendix to Mixed Modeling for Irregularly Sampled and Correlated Functional Data: Speech Science Spplications
|
|
|
- Percival Tucker
- 5 years ago
- Views:
Transcription
1 Online Appendix to Mixed Modeling for Irregularly Sampled and Correlated Functional Data: Speech Science Spplications Marianne Pouplier, Jona Cederbaum, Philip Hoole, Stefania Marin, Sonja Greven R Syntax and Plotting Commands for Example Datasets This Online Appendix illustrates how the sparseflmm package is used to run the case studies discussed in the paper. Plotting commands are exemplified in order to help the reader understand the structure of the output. A generally adaptable plotting function is provided for one of the datasets on the basis of which the reader should, mutatis mutandis, be able to generate plots for their own data in many cases. We provide with this Appendix the input data for each case study and a results file so that the reader can examine the output without having to run the models. Installation The R package sparseflmm can be downloaded from CRAN. For installation, R version or later is required, as are the following packages: mgcv (>= ) data.table (>= 1.9-5) refund (>= ) First load the package: require(sparseflmm) Here is how the citation information for the package can be retrieved: citation("sparseflmm") Romanian Vowel Sequences Model Specification The Romanian vowel F2 data were analysed with a model specifying a single covariate and three functional random effects (Subject (B), Item (C), Subject-Item-Repetition (E)). 1
2 The input to the model is an R data table. Any data frame can be easily transformed to a data.table by applying the function data.table() on an existing data frame. The advantage of data table is that it is faster and more memory efficient. The data table must contain the following columns: n_long (integer): unique identification number for each curve. In this dataset n_long ranges from 1:163, since there are 163 curves for analysis. subject_long (integer): unique identification number for each level of the first functional random effect (term B in equation (1) of the main paper), here Subject. Since we have 5 speakers, subject_long ranges from 1:5 word_long (integer): unique identification number for each level of the second functional random effect (term C in equation (1) of the main paper). This study has 6 stimulus items (3 diphthong, 3 hiatus items), therefore word_long takes on the range 1:6 combi_long (integer): repetition of each subject_long word_long combination. Here this ranges from 1:6 since each speaker recorded each stimulus item six times. y_vec (numeric): F2 value of each sample t (numeric): the normalized time points at which the F2 values were sampled covariate.1 (numeric): covariate values, here dummy coded levels 0 = diphthong, 1 = hiatus Note that the data table has to be in long format, in which each row represents a single data point. This means that, except for the coordinates of each sample, the coding is redundant within a given curve. The first step is to load the data table into the workspace. In our case it is called RoDiphF2: load("rodiphf2.rdata") head(rodiphf2) ## n_long subject_long word_long combi_long y_vec t covariate.1 ## 1: ## 2: ## 3: ## 4: ## 5: ## 6: When calling sparseflmm the parameters for which there are no defaults or for which we want to depart from the default settings have to be specified (see?sparseflmm for information on defaults). The data table with the input data is given to the function as variable curve_info: curve_info = RoDiphF2 We have a single covariate and specify the covariate form. The covariate form needs one entry for each covariate (here, one). Since the covariate is discrete (dummy coded), the only 2
3 option is by covariate = TRUE num_covariates = 1 covariate_form = by We have no interaction, hence interaction = FALSE We specify in bf_covs the number of basis functions for each functional random effect (here, three: Subject, Item, Subject-Item-Repetition). It is important to choose a sufficiently high number of basis functions. Which number can be considered sufficient depends on the complexity and smoothness of the curves. Since during the estimation the curves are represented as a linear combination of the given number of basis functions, this number basically determines the amount of detail retained from the original curves. While a high number is desirable (over-fitting is avoided by penalty terms) it is computationally very costly. One way to determine the appropriate number of basis functions is to successively increase the number of basis functions until the output stabilizes. The term m_covs is a list that specifies for each functional random intercept the order of the spline and of the penalty for covariance estimation (here, cubic B-Splines which corresponds to order = 2, and the third derivative). bf_covs = c(10, 10, 10) m_covs = list(c(2,3), c(2,3), c(2,3)) For the number of basis functions for the mean (bf_mean) we use the default of 8 and thus no further specification in the call to the function is needed for this parameter. Generally, as for the functional random effects, a higher number of basis functions is advantageous in the estimation since more detailed information on curve shape is preserved. Over-fitting is again avoided due to the penalty term. A high number of basis functions is, however, computationally very costly (see main paper for more discussion and references). Using the third derivative (jerk) for the penalty of the covariance estimation can be motivated additionally for speech data under the consideration that jerk minimization is commonly considered to be an important factor in movement optimization. Since we want confidence bands to be computed, we require functional additive mixed modelling (FAMM) to be used use_famm = TRUE Here is the call to the function: results <- sparseflmm(curve_info = RoDiphF2, covariate = TRUE, num_covariates = 1, covariate_form = "by", interaction = FALSE, bf_covs = c(10,10,10), m_covs = list(c(2,3), c(2,3), c(2,3)), use_famm = TRUE) This dataset is small and the analysis can be run on an ordinary desktop computer. 3
4 Output Effects Plots For present purposes we first load a file with the results so that we can look at the results without having to run the model. load("resultsrodiphf2.rdata") For plotting the reference mean, we extract the effect of the reference mean from the fpc_famm estimation and add it to the intercept. This recreates Figure 2 of the paper. intercept <- results$fpc_famm_hat_tri_constr$intercept y_mean <- results$fpc_famm_hat_tri_constr$famm_cb_mean$value se_mean <- results$fpc_famm_hat_tri_constr$famm_cb_mean$se my_grid <- results$my_grid plot(my_grid, y_mean+intercept, t = "l", lwd=3,xlab = "normalized time (t)", ylab = "delta Hz", ylim = c(1000,2000), col = "red", cex.axis = 1.2, main = expression(paste("reference Mean ", f[0](t))),cex.main = 1.2,cex.lab = 1.2) #add confidence bands to the plot lines(x = my_grid, y = (y_mean+2*se_mean+intercept), lty = 2, lwd = 3, col = "blue") lines(x = my_grid, y = (y_mean-2*se_mean+intercept), lty = 2, lwd = 3, col = "blue") Reference Mean f 0 (t) delta Hz normalized time (t) 4
5 In the same way we plot the effect of the covariate: y_cov1<-results$fpc_famm_hat_tri_constr$famm_cb_covariate.1$value se_cov1<-results$fpc_famm_hat_tri_constr$famm_cb_covariate.1$se plot(my_grid,y_cov1, t = "l", lwd = 3, xlab = " normalized time (t)", ylab = "delta Hz", ylim = c(-200,800), col = "red",cex.axis = 1.2, main = expression(paste("status ", f[1](t))), cex.main = 1.2, cex.lab = 1.2) #confidence bands lines(x = my_grid, y = (y_cov1+2*se_cov1), lty = 2, lwd = 3, col = "blue") lines(x = my_grid, y = (y_cov1-2*se_cov1), lty = 2, lwd = 3, col = "blue") #reference zero line abline(h = 0, lty = 3) Status f 1 (t) delta Hz normalized time (t) Summed Effect Curves To obtain the summed effect curves we sum the intercept, the mean, and the covariate effect. # define colors and scaling of the y-axis cols <- rainbow(12) miny <
6 maxy < # sum the intercept, mean and intercept, mean, covariate effect none <- intercept+y_mean Status <- intercept+y_mean+y_cov1 #plot first the reference mean and then the covariate effect plot(my_grid, none, col = cols[1], lty = 1, lwd = 4, t = "l", ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n") plot(my_grid, Status, col = cols[10], lty = 4, lwd = 4, ylim = c(miny, maxy), ylab = "delta Hz", xlab = "normalized time", main = paste0("summed Effect Curves"), t = "l", cex.axis = 1.2, cex.main = 1.2, font.main = 1, cex.lab = 1.2) #zero reference line abline(h = 0, lty = 3) # legend text leg.text <- c(expression(paste(f[0](t))), expression(paste(f[0](t)+f[1](t)))) legend("bottomleft", legend = leg.text, bty = "n", col = c(cols[1], cols[10]), lty = c(1,4), lwd = 4,cex = 1.2) Summed Effect Curves delta Hz f 0 (t) f 0 (t) + f 1 (t) normalized time 6
7 Variance Decomposition For variance decomposition, results$fpc_hat_tri_constr has the information needed to obtain the number of FPCs chosen for each functional random effect, the percent variance explained by each FPC, and the weights associated with each chosen FPC. results$fpc_hat_tri_constr$n_b/c/e contains the number of FPCs chosen for each functional random effect, here 3 for B (Speaker), 1 for C (Item), 3 for E (Speaker-Item-Repetition). The estimated total variance explained can be found in results$fpc_hat_tri_constr$var_explained ## [1] To calculate the amount of variance explained by each FPC we need the estimated total variance totvar_hat <- results$fpc_hat_tri_constr$total_var print(totvar_hat) ## [1] as well as the estimated eigenvalues for each functional random effect results$fpc_hat_tri_constr$nu_b/c/e_hat The estimated variance accounted for by the functional random effect Speaker (B) in total is then obtained like this: sum(results$fpc_hat_tri_constr$nu_b_hat)/totvar_hat*100 ## [1] This renders the variance accounted for by each of the FPCs selected for Speaker: results$fpc_hat_tri_constr$nu_b_hat/totvar_hat*100 ## [1] Accordingly, the estimated eigenvalue for the functional random effect C (Item) is results$fpc_hat_tri_constr$nu_c_hat/totvar_hat*100 ## [1] The variance accounted for by random error results$cov_hat_tri_constr$sigmasq/totvar_hat*100 ## [1] To obtain plots as given in Figures 3 and 4 of the paper, we need to calculate twice the square root of the relevant eigenvalue and retrieve the estimated eigenfunctions (FPCs) for 7
8 the given functional random effect. Here we illustrate this for Speaker (B), in correspondence to Figure 3 of the paper. mult <- sqrt(results$fpc_hat_tri_constr$nu_b_hat[1])*2 phi_b_hat_grid <- results$fpc_hat_tri_constr$phi_b_hat_grid The global mean is obtained by setting the covariates to their mean (0.5): intercept <- results$fpc_famm_hat_tri_constr$intercept gmean <- intercept+y_mean+y_cov1*0.5 The left part of Figure 3 of the paper which gives FPC1 chosen for functional random effect Speaker (B) is then obtained as follows: #percent variance accounted for (for legend) exvarb_sep <- results$fpc_hat_tri_constr$nu_b_hat/totvar_hat*100 # set margins so that the ylabel is not cut off par(mar = c (5, 5, 4, 2)) #plot the global mean plot(my_grid, gmean, type = "l", ylim = c(1200,2400), xlab = "normalized time",ylab = expression(paste(phi[1]^b, "(t)")), cex.axis = 1.2) #plot the suitable multiple (twice the square root of the eigenvalue) points(my_grid, gmean+mult*phi_b_hat_grid[, 1], pch = "+", t = "p") points(my_grid, gmean-mult*phi_b_hat_grid[, 1], pch = "-", t = "p") #legend text legend("topright", legend = paste0("explains ", round(exvarb_sep[1], 2), "%"), bty = "n", cex = 1.2) 8
9 φ 1 B (t) explains 37.64% normalized time To get each speaker s random intercept curve (right part of Figure 3), we proceed as follows: selected_use <- 1:5 # plot means for all 5 speakers color_use <- rainbow(length(selected_use)) #define colors #retrieve the random intercept curve per speaker B_hat <- results$fpc_famm_hat_tri_constr$famm_predict_b #add the random intercept curves to the global mean B_hat_plus_mean <- t(t(b_hat)+gmean) #plot the global mean plot(my_grid, gmean, ylim = c(1200,2400), lty = 2, lwd = 3, t = "l", xlab = "", ylab="") #plot the estimated random effects curve for each speaker for (ii in selected_use){ plot(my_grid, t(b_hat_plus_mean[ii,]), t = "l", lty = ii, lwd = 3, ylim = c(1200,2400), col = color_use[ii], xlab = "normalized time", ylab = expression(paste(mu(t)+b[i], "(t)"))) } 9
10 #legend text legend("topright",legend = (c("global mean", selected_use)), text.col = c("#000000",color_use), bty = "n", lty = c(2,1:5), col = c("#000000",color_use),lwd = 3) µ(t) + B i (t) global mean normalized time All of the above steps are summarized in the plotting functions plot_resultsrodiph.r and plot_summed_effectsrodiph.r provided as part of this Appendix. The functions are specific to the Romanian dataset but they can straightfowardly be adapted to the other example datasets and other model outputs. EPG Data of German Fricative Sequences Model Specification The EPG data were analysed using a model with two covariates and their interaction. Again this is a model with crossed functional random effects for Subject, Item, and Subject-Item- Repetition. 10
11 load("epgdata.rdata") Curve_info is set to the data table with the input data: curve_info = EPGdata We have two covariates. Form needs to be specified for each covariate. covariate = TRUE num_covariates = 2 covariate_form = rep( by,2) This time we have an interaction, hence we specify the symmetric interaction matrix with as many rows and columns as there are covariates: interaction = TRUE which_interaction = matrix(c(false, TRUE, TRUE, FALSE), byrow = TRUE, nrow = 2, ncol = 2) We specify the number of basis functions and the penalty in bf_covs and m_covs in the same way as for the Romanian vowel model since there are three functional random effects: bf_covs = c(10,10,10) m_covs = list(c(2,3), c(2,3), c(2,3)) Since we want confidence bands to be computed, we require FAMM to be used: use_famm = TRUE Here is the call to the function: results <- sparseflmm(curve_info = EPGData, covariate = TRUE, num_covariates = 2, covariate_form = rep("by",2), interaction = TRUE, which_interaction = matrix(c(false, TRUE,TRUE,FALSE), byrow = TRUE,nrow = 2,ncol = 2), bf_covs = c(10,10,10), m_covs = list(c(2,3),c(2,3),c(2,3)), use_famm = TRUE) Output We again provide a results file so that plots can be generated without having to run the model. load("resultsepgdata.rdata") 11
12 Effects Plots Plotting the effects is analogous to the previous example, we only add plotting commands for the second covariate and the interaction. #retrieve the intercept, reference mean and confidence bands intercept <- results$fpc_famm_hat_tri_constr$intercept y_mean <- results$fpc_famm_hat_tri_constr$famm_cb_mean$value se_mean <- results$fpc_famm_hat_tri_constr$famm_cb_mean$se my_grid <- results$my_grid plot(my_grid, y_mean+intercept, lwd = 3,t = "l", xlab = "normalized time (t)", ylab = "delta index", ylim = c(-1,1), col = "red", cex.axis = 1.2, main = expression(paste("reference Mean ", f[0](t))), cex.main = 1.2, cex.lab = 1.2) #confidence bands lines(x = my_grid, y = (y_mean+2*se_mean+intercept), lty = 2, lwd = 3,col = "blue") lines(x = my_grid, y = (y_mean-2*se_mean+intercept), lty = 2, lwd = 3,col = "blue") # zero reference line abline(h = 0, lty = 3) Reference Mean f 0 (t) delta index normalized time (t) 12
13 The plot for covariate.1 (C1): y_cov1 <- results$fpc_famm_hat_tri_constr$famm_cb_covariate.1$value se_cov1 <- results$fpc_famm_hat_tri_constr$famm_cb_covariate.1$se plot(my_grid, y_cov1, t = "l",lwd = 3, xlab = " normalized time (t)", ylab = "delta index", ylim = c(-1,1), col = "red", cex.axis = 1.2, main = expression(paste("c1 ", f[1](t))), cex.main = 1.2, cex.lab = 1.2) lines(x = my_grid, y = (y_cov1+2*se_cov1), lty = 2, lwd = 3, col = "blue") lines(x = my_grid, y = (y_cov1-2*se_cov1), lty = 2, lwd = 3, col = "blue") abline(h = 0, lty = 3) C1 f 1 (t) delta index normalized time (t) The plot for covariate.2 (Stress): y_cov2 <- results$fpc_famm_hat_tri_constr$famm_cb_covariate.2$value se_cov2 <- results$fpc_famm_hat_tri_constr$famm_cb_covariate.2$se plot(my_grid, y_cov2, t = "l",lwd = 3, xlab = " normalized time (t)", ylab = "delta index", ylim = c(-1,1), col = "red", cex.axis = 1.2, main = expression(paste("stress ", f[2](t))), cex.main = 1.2, cex.lab = 1.2) lines(x = my_grid, y = (y_cov2+2*se_cov2), lty = 2, lwd = 3, col = "blue") lines(x = my_grid, y = (y_cov2-2*se_cov2), lty = 2, lwd = 3, col = "blue") abline(h = 0, lty = 3) 13
14 Stress f 2 (t) delta index normalized time (t) The plot for the C1 * Stress Interaction is obtained analogously: y_inter_1_2 <- results$fpc_famm_hat_tri_constr$famm_cb_inter_1_2$value se_inter_1_2 <- results$fpc_famm_hat_tri_constr$famm_cb_inter_1_2$se plot(my_grid, y_inter_1_2, t = "l", lwd = 3, xlab = " normalized time (t)", ylab = "delta index", ylim = c(-1,1), col = "red", cex.axis = 1.2, main = expression(paste("c1 * Stress ", f[3](t))), cex.main = 1.2, cex.lab = 1.2) lines(x = my_grid, y = (y_inter_1_2+2*se_inter_1_2), lty = 2,lwd = 3,col = "blue") lines(x = my_grid, y = (y_inter_1_2-2*se_inter_1_2), lty = 2,lwd = 3,col = "blue") abline(h = 0, lty = 3) 14
15 C1 * Stress f 3 (t) delta index normalized time (t) Summed Effects Again we proceed in analogy to the Romanian vowel data, only adding a second covariate and the interaction. #define colors and scaling of y-axis cols <- rainbow(12) miny <- -1 maxy <- 1 #define the summed effects none <- intercept+y_mean C1 <- intercept+y_mean+y_cov1 Stress <- intercept+y_mean+y_cov2 Stress_C1 <- intercept+y_mean+y_cov1+y_cov2+y_inter_1_2 plot(my_grid, none, col = cols[1], lty = 1,lwd = 4,t = "l", ylim = c(miny, maxy), ylab = "", xlab = "", xaxt="n", yaxt = "n") plot(my_grid, C1, col = cols[8], lty = 4, lwd = 4,t = "l", ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n") plot(my_grid, Stress, col = cols[2], lty = 5,lwd = 4, t = "l", ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n") 15
16 plot(my_grid, Stress_C1, col = cols[9], lty = 3,lwd = 4, ylim = c(miny, maxy), ylab = "delta index",xlab = "normalized time", main = expression(paste("summed Effects: C1 ", f[1](t), ", Stress ", f[2](t), ", and Their Interaction ", f[3](t))), t = "l", cex.axis=1.2, cex.main = 1.2, cex.lab = 1.2) abline(h = 0,lty = 3) leg.text <- c(expression(paste(f[0](t))), expression(paste(f[0](t)+f[1](t))), expression(paste(f[0](t)+f[2](t))), expression(paste(f[0](t)+f[1](t)+f[2](t)+f[3](t)))) legend("topright", legend = leg.text, bty = "n", col = c(cols[1], cols[8], cols[2], cols[9]), lty = c(1,4,5,9,2,1,1,1), lwd = c(4,4,4,4)) Summed Effects: C1 f 1 (t), Stress f 2 (t), and Their Interaction f 3 (t) delta index f 0 (t) f 0 (t) + f 1 (t) f 0 (t) + f 2 (t) f 0 (t) + f 1 (t) + f 2 (t) + f 3 (t) normalized time 16
17 Variance Decomposition The procedures for plotting variance decomposition are analogous to the Romanian vowel example and will not be repeated here. We only point out that in the case of two covariates and an interaction the global mean is computed as follows: intercept <- results$fpc_famm_hat_tri_constr$intercept y_mean <- results$fpc_famm_hat_tri_constr$famm_cb_mean$value y_cov1 <- results$fpc_famm_hat_tri_constr$famm_cb_covariate.1$value y_cov2 <- results$fpc_famm_hat_tri_constr$famm_cb_covariate.2$value y_inter_1_2 <- results$fpc_famm_hat_tri_constr$famm_cb_inter_1_2$value gmean <- intercept+y_mean+y_cov1*0.5+y_cov2*0.5+y_inter_1_2*0.25 This is also implemented in the plot_results function provided with the Romanian data in case an interaction is specified. Frequency Effects in Articulography Data Model Specification For the Russian articulography data, we use a model with a functional random intercept for Subject and for Subject-Repetition. There is no functional random effect for Item, since each frequency is only represented by a single syllable. load("freqdata.rdata") First we specify the input data table as curve_info curve_info = FreqData The default setting is for crossed functional random effects. Since we only have a functional random effect for Speaker here, we specify use_ri = TRUE There is a single covariate and no interaction. Note that the continuous covariate has to be centered prior to running the model. covariate = TRUE num_covariate = 1 covariate_form = by interaction = FALSE The number of basis functions is specified for the functional random effects: bf_covs = c(10, 10) m_covs = list(c(2, 3), c(2, 3)) To obtain confidence bands, set 17
18 use_famm = TRUE Since this is a large dataset, we use two cores to parallelize the auto-covariance and FAMM estimation: para_estim_cov = TRUE para_estim_cov_nc = 2 para_estim_famm = TRUE para_estim_famm_nc = 2 results <- sparseflmm(curve_info = FreqData, use_ri = TRUE, covariate = TRUE, num_covariate = 1, covariate_form = "by", interaction = FALSE, bf_covs = c(10, 10), m_covs = list(c(2, 3), c(2, 3)), use_famm = TRUE, para_estim_cov = TRUE, para_estim_cov_nc = 2, para_estim_famm = TRUE, para_estim_famm_nc = 2) Output The effects plots are generated as before and the plotting commands are not repeated here. Specific to this dataset is, besides the use of only two functional random effects, the use of a continuous covariate. This means that in order to understand the estimated effect of frequency on the kinematics of C2, it is advisable to plot the summed effects for a few selected frequency values (alternatively, for instance the quantiles could be used for plotting). This is done by adding to the reference mean the estimated curve of covariate.1 (Frequency) multiplied by the given centered frequency value of interest. Again we load the provided results file load("resultsfreqdata.rdata") The centered frequency values are retrieved from the input data table. fs <- unique(freqdata$covariate.1) fs <- sort(fs) As in the previous cases, we retrieve the intercept, the reference mean, and the covariate.1 (Frequency) effect. intercept <- results$fpc_famm_hat_tri_constr$intercept my_grid <- results$my_grid y_mean <- results$fpc_famm_hat_tri_constr$famm_cb_mean$value y_cov1 <- results$fpc_famm_hat_tri_constr$famm_cb_covariate.1$value # 5 frequencies are chosen for the summed effects plot freq1 <- intercept+y_mean+y_cov1*fs[1] #lowest frequency (/mx/, freq=6.0) freq4 <- intercept+y_mean+y_cov1*fs[4] 18
19 freq8 <- intercept+y_mean+y_cov1*fs[8] freq9 <- intercept+y_mean+y_cov1*fs[9] freq12 <- intercept+y_mean+y_cov1*fs[12] #highest frequency (/gl/, freq=12.7) none <- intercept+y_mean miny <- -2 maxy <- 2 cols <- rainbow(12) plot(my_grid, my_grid, t = "n", ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt="n") plot(my_grid, none, col = "#000000", lty = 1,t = "l", ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n",lwd = 4) plot(my_grid, freq1, col = cols[2], lty = 2, ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n", t = "l",lwd = 4) plot(my_grid, freq4, col = cols[6], lty = 3, ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n", t = "l",lwd = 4) plot(my_grid, freq8, col = cols[7], lty = 4, ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n", t = "l",lwd = 4) plot(my_grid, freq9, col = cols[9], lty = 5, ylim = c(miny, maxy), ylab = "", xlab = "", xaxt = "n", yaxt = "n", t = "l",lwd=4) plot(my_grid, freq12, col = cols[12], lty = 6, ylim = c(miny, maxy), ylab = "delta position", xlab = "normalized time", t = "l",lwd=4, main = paste0("summed Effect Curves"), cex.axis = 1.2, cex.main = 1.2) leg.text <- c(expression(paste(f[0](t))), expression(paste(f[0](t)+f[1](t),"*",freq1)), expression(paste(f[0](t)+f[1](t),"*",freq4)), expression(paste(f[0](t)+f[1](t),"*",freq8)), expression(paste(f[0](t)+f[1](t),"*",freq9)), expression(paste(f[0](t)+f[1](t),"*",freq12))) legend("topleft", legend = leg.text, bty = "n", col = c("#000000", cols[2], cols[6], cols[7], cols[9], cols[12]), lty = 1:6, lwd = 4) 19
20 abline(h = 0, lty = 3) Summed Effect Curves delta position f 0 (t) f 0 (t) + f 1 (t)*freq1 f 0 (t) + f 1 (t)*freq4 f 0 (t) + f 1 (t)*freq8 f 0 (t) + f 1 (t)*freq9 f 0 (t) + f 1 (t)*freq normalized time 20
Package sparseflmm. September 23, 2017
 Type Package Package sparseflmm September 23, 2017 Title Functional Linear Mixed Models for Irregularly or Sparsely Sampled Data Version 0.2.0 Author Jona Cederbaum Maintainer Jona Cederbaum
Type Package Package sparseflmm September 23, 2017 Title Functional Linear Mixed Models for Irregularly or Sparsely Sampled Data Version 0.2.0 Author Jona Cederbaum Maintainer Jona Cederbaum
Canadian climate: function-on-function regression
 Canadian climate: function-on-function regression Sarah Brockhaus Institut für Statistik, Ludwig-Maximilians-Universität München, Ludwigstraße 33, D-0539 München, Germany. The analysis is based on the
Canadian climate: function-on-function regression Sarah Brockhaus Institut für Statistik, Ludwig-Maximilians-Universität München, Ludwigstraße 33, D-0539 München, Germany. The analysis is based on the
Package denseflmm. R topics documented: March 17, 2017
 Package denseflmm March 17, 2017 Type Package Title Functional Linear Mixed Models for Densely Sampled Data Version 0.1.0 Author Sonja Greven, Jona Cederbaum Maintainer Jona Cederbaum
Package denseflmm March 17, 2017 Type Package Title Functional Linear Mixed Models for Densely Sampled Data Version 0.1.0 Author Sonja Greven, Jona Cederbaum Maintainer Jona Cederbaum
Functional Linear Mixed Models for. Irregularly or Sparsely Sampled Data
 Functional Linear Mixed Models for Irregularly or Sparsely Sampled Data Jona Cederbaum 1, Marianne Pouplier 2, Phil Hoole 2, and Sonja Greven 1 arxiv:150801686v1 [statme] 7 Aug 2015 1 Department of Statistics,
Functional Linear Mixed Models for Irregularly or Sparsely Sampled Data Jona Cederbaum 1, Marianne Pouplier 2, Phil Hoole 2, and Sonja Greven 1 arxiv:150801686v1 [statme] 7 Aug 2015 1 Department of Statistics,
Lecture 5 : The Poisson Distribution
 Lecture 5 : The Poisson Distribution Jonathan Marchini November 5, 2004 1 Introduction Many experimental situations occur in which we observe the counts of events within a set unit of time, area, volume,
Lecture 5 : The Poisson Distribution Jonathan Marchini November 5, 2004 1 Introduction Many experimental situations occur in which we observe the counts of events within a set unit of time, area, volume,
Package interspread. September 7, Index 11. InterSpread Plus: summary information
 Package interspread September 7, 2012 Version 0.2-2 Date 2012-09-07 Title Functions for analysing InterSpread Plus simulation output Author Mark Stevenson A package for analysing
Package interspread September 7, 2012 Version 0.2-2 Date 2012-09-07 Title Functions for analysing InterSpread Plus simulation output Author Mark Stevenson A package for analysing
Metric Predicted Variable on Two Groups
 Metric Predicted Variable on Two Groups Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Goals
Metric Predicted Variable on Two Groups Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Goals
Package face. January 19, 2018
 Package face January 19, 2018 Title Fast Covariance Estimation for Sparse Functional Data Version 0.1-4 Author Luo Xiao [aut,cre], Cai Li [aut], William Checkley [aut], Ciprian Crainiceanu [aut] Maintainer
Package face January 19, 2018 Title Fast Covariance Estimation for Sparse Functional Data Version 0.1-4 Author Luo Xiao [aut,cre], Cai Li [aut], William Checkley [aut], Ciprian Crainiceanu [aut] Maintainer
A Handbook of Statistical Analyses Using R 3rd Edition. Torsten Hothorn and Brian S. Everitt
 A Handbook of Statistical Analyses Using R 3rd Edition Torsten Hothorn and Brian S. Everitt CHAPTER 6 Simple and Multiple Linear Regression: How Old is the Universe and Cloud Seeding 6.1 Introduction?
A Handbook of Statistical Analyses Using R 3rd Edition Torsten Hothorn and Brian S. Everitt CHAPTER 6 Simple and Multiple Linear Regression: How Old is the Universe and Cloud Seeding 6.1 Introduction?
Package clustergeneration
 Version 1.3.4 Date 2015-02-18 Package clustergeneration February 19, 2015 Title Random Cluster Generation (with Specified Degree of Separation) Author Weiliang Qiu , Harry Joe
Version 1.3.4 Date 2015-02-18 Package clustergeneration February 19, 2015 Title Random Cluster Generation (with Specified Degree of Separation) Author Weiliang Qiu , Harry Joe
Package rnmf. February 20, 2015
 Type Package Title Robust Nonnegative Matrix Factorization Package rnmf February 20, 2015 An implementation of robust nonnegative matrix factorization (rnmf). The rnmf algorithm decomposes a nonnegative
Type Package Title Robust Nonnegative Matrix Factorization Package rnmf February 20, 2015 An implementation of robust nonnegative matrix factorization (rnmf). The rnmf algorithm decomposes a nonnegative
Package ForwardSearch
 Package ForwardSearch February 19, 2015 Type Package Title Forward Search using asymptotic theory Version 1.0 Date 2014-09-10 Author Bent Nielsen Maintainer Bent Nielsen
Package ForwardSearch February 19, 2015 Type Package Title Forward Search using asymptotic theory Version 1.0 Date 2014-09-10 Author Bent Nielsen Maintainer Bent Nielsen
Package msir. R topics documented: April 7, Type Package Version Date Title Model-Based Sliced Inverse Regression
 Type Package Version 1.3.1 Date 2016-04-07 Title Model-Based Sliced Inverse Regression Package April 7, 2016 An R package for dimension reduction based on finite Gaussian mixture modeling of inverse regression.
Type Package Version 1.3.1 Date 2016-04-07 Title Model-Based Sliced Inverse Regression Package April 7, 2016 An R package for dimension reduction based on finite Gaussian mixture modeling of inverse regression.
Measurement, Scaling, and Dimensional Analysis Summer 2017 METRIC MDS IN R
 Measurement, Scaling, and Dimensional Analysis Summer 2017 Bill Jacoby METRIC MDS IN R This handout shows the contents of an R session that carries out a metric multidimensional scaling analysis of the
Measurement, Scaling, and Dimensional Analysis Summer 2017 Bill Jacoby METRIC MDS IN R This handout shows the contents of an R session that carries out a metric multidimensional scaling analysis of the
Package CorrMixed. R topics documented: August 4, Type Package
 Type Package Package CorrMixed August 4, 2016 Title Estimate Correlations Between Repeatedly Measured Endpoints (E.g., Reliability) Based on Linear Mixed-Effects Models Version 0.1-13 Date 2015-03-08 Author
Type Package Package CorrMixed August 4, 2016 Title Estimate Correlations Between Repeatedly Measured Endpoints (E.g., Reliability) Based on Linear Mixed-Effects Models Version 0.1-13 Date 2015-03-08 Author
R-companion to: Estimation of the Thurstonian model for the 2-AC protocol
 R-companion to: Estimation of the Thurstonian model for the 2-AC protocol Rune Haubo Bojesen Christensen, Hye-Seong Lee & Per Bruun Brockhoff August 24, 2017 This document describes how the examples in
R-companion to: Estimation of the Thurstonian model for the 2-AC protocol Rune Haubo Bojesen Christensen, Hye-Seong Lee & Per Bruun Brockhoff August 24, 2017 This document describes how the examples in
Modern Regression HW #6 Solutions
 36-401 Modern Regression HW #6 Solutions Problem 1 [32 points] (a) (4 pts.) DUE: 10/27/2017 at 3PM Given : Chick 50 150 300 50 150 300 50 150 300 50 150 300 Weight 50 150 300 50 150 300 50 150 300 Figure
36-401 Modern Regression HW #6 Solutions Problem 1 [32 points] (a) (4 pts.) DUE: 10/27/2017 at 3PM Given : Chick 50 150 300 50 150 300 50 150 300 50 150 300 Weight 50 150 300 50 150 300 50 150 300 Figure
Package idmtpreg. February 27, 2018
 Type Package Package idmtpreg February 27, 2018 Title Regression Model for Progressive Illness Death Data Version 1.1 Date 2018-02-23 Author Leyla Azarang and Manuel Oviedo de la Fuente Maintainer Leyla
Type Package Package idmtpreg February 27, 2018 Title Regression Model for Progressive Illness Death Data Version 1.1 Date 2018-02-23 Author Leyla Azarang and Manuel Oviedo de la Fuente Maintainer Leyla
Problems from Chapter 3 of Shumway and Stoffer s Book
 UNIVERSITY OF UTAH GUIDED READING TIME SERIES Problems from Chapter 3 of Shumway and Stoffer s Book Author: Curtis MILLER Supervisor: Prof. Lajos HORVATH November 10, 2015 UNIVERSITY OF UTAH DEPARTMENT
UNIVERSITY OF UTAH GUIDED READING TIME SERIES Problems from Chapter 3 of Shumway and Stoffer s Book Author: Curtis MILLER Supervisor: Prof. Lajos HORVATH November 10, 2015 UNIVERSITY OF UTAH DEPARTMENT
Dispersion of a flow that passes a dynamic structure. 5*t 5*t. Time
 > # PROJECT > # ======= > # Principles of Modelling and Simulation in Epidemiology > # Laboratory exercise 1 > # > # DESCRIPTION > # =========== > # Suggestions of solutions > > # THOR AND DATE > # Andreas
> # PROJECT > # ======= > # Principles of Modelling and Simulation in Epidemiology > # Laboratory exercise 1 > # > # DESCRIPTION > # =========== > # Suggestions of solutions > > # THOR AND DATE > # Andreas
A Handbook of Statistical Analyses Using R. Brian S. Everitt and Torsten Hothorn
 A Handbook of Statistical Analyses Using R Brian S. Everitt and Torsten Hothorn CHAPTER 5 Multiple Linear Regression: Cloud Seeding 5.1 Introduction 5.2 Multiple Linear Regression 5.3 Analysis Using R
A Handbook of Statistical Analyses Using R Brian S. Everitt and Torsten Hothorn CHAPTER 5 Multiple Linear Regression: Cloud Seeding 5.1 Introduction 5.2 Multiple Linear Regression 5.3 Analysis Using R
Matematisk statistik allmän kurs, MASA01:A, HT-15 Laborationer
 Lunds universitet Matematikcentrum Matematisk statistik Matematisk statistik allmän kurs, MASA01:A, HT-15 Laborationer General information on labs During the rst half of the course MASA01 we will have
Lunds universitet Matematikcentrum Matematisk statistik Matematisk statistik allmän kurs, MASA01:A, HT-15 Laborationer General information on labs During the rst half of the course MASA01 we will have
Package CEC. R topics documented: August 29, Title Cross-Entropy Clustering Version Date
 Title Cross-Entropy Clustering Version 0.9.4 Date 2016-04-23 Package CEC August 29, 2016 Author Konrad Kamieniecki [aut, cre], Przemyslaw Spurek [ctb] Maintainer Konrad Kamieniecki
Title Cross-Entropy Clustering Version 0.9.4 Date 2016-04-23 Package CEC August 29, 2016 Author Konrad Kamieniecki [aut, cre], Przemyslaw Spurek [ctb] Maintainer Konrad Kamieniecki
Package leiv. R topics documented: February 20, Version Type Package
 Version 2.0-7 Type Package Package leiv February 20, 2015 Title Bivariate Linear Errors-In-Variables Estimation Date 2015-01-11 Maintainer David Leonard Depends R (>= 2.9.0)
Version 2.0-7 Type Package Package leiv February 20, 2015 Title Bivariate Linear Errors-In-Variables Estimation Date 2015-01-11 Maintainer David Leonard Depends R (>= 2.9.0)
Jian WANG, PhD. Room A115 College of Fishery and Life Science Shanghai Ocean University
 Jian WANG, PhD j_wang@shou.edu.cn Room A115 College of Fishery and Life Science Shanghai Ocean University Contents 1. Introduction to R 2. Data sets 3. Introductory Statistical Principles 4. Sampling and
Jian WANG, PhD j_wang@shou.edu.cn Room A115 College of Fishery and Life Science Shanghai Ocean University Contents 1. Introduction to R 2. Data sets 3. Introductory Statistical Principles 4. Sampling and
Package gma. September 19, 2017
 Type Package Title Granger Mediation Analysis Version 1.0 Date 2018-08-23 Package gma September 19, 2017 Author Yi Zhao , Xi Luo Maintainer Yi Zhao
Type Package Title Granger Mediation Analysis Version 1.0 Date 2018-08-23 Package gma September 19, 2017 Author Yi Zhao , Xi Luo Maintainer Yi Zhao
Chapter 4 Exercises 1. Data Analysis & Graphics Using R Solutions to Exercises (December 11, 2006)
 Chapter 4 Exercises 1 Data Analysis & Graphics Using R Solutions to Exercises (December 11, 2006) Preliminaries > library(daag) Exercise 2 Draw graphs that show, for degrees of freedom between 1 and 100,
Chapter 4 Exercises 1 Data Analysis & Graphics Using R Solutions to Exercises (December 11, 2006) Preliminaries > library(daag) Exercise 2 Draw graphs that show, for degrees of freedom between 1 and 100,
Metric Predicted Variable With One Nominal Predictor Variable
 Metric Predicted Variable With One Nominal Predictor Variable Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more
Metric Predicted Variable With One Nominal Predictor Variable Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more
Lab 9: An Introduction to Wavelets
 Lab 9: An Introduction to Wavelets June 5, 2003 In this lab, we introduce some basic concepts of wavelet theory and present the essentials required for possible applications of wavelet decompositions for
Lab 9: An Introduction to Wavelets June 5, 2003 In this lab, we introduce some basic concepts of wavelet theory and present the essentials required for possible applications of wavelet decompositions for
The coxvc_1-1-1 package
 Appendix A The coxvc_1-1-1 package A.1 Introduction The coxvc_1-1-1 package is a set of functions for survival analysis that run under R2.1.1 [81]. This package contains a set of routines to fit Cox models
Appendix A The coxvc_1-1-1 package A.1 Introduction The coxvc_1-1-1 package is a set of functions for survival analysis that run under R2.1.1 [81]. This package contains a set of routines to fit Cox models
Package hds. December 31, 2016
 Type Package Version 0.8.1 Title Hazard Discrimination Summary Package hds December 31, 2016 Functions for calculating the hazard discrimination summary and its standard errors, as described in Liang and
Type Package Version 0.8.1 Title Hazard Discrimination Summary Package hds December 31, 2016 Functions for calculating the hazard discrimination summary and its standard errors, as described in Liang and
Package depth.plot. December 20, 2015
 Package depth.plot December 20, 2015 Type Package Title Multivariate Analogy of Quantiles Version 0.1 Date 2015-12-19 Maintainer Could be used to obtain spatial depths, spatial ranks and outliers of multivariate
Package depth.plot December 20, 2015 Type Package Title Multivariate Analogy of Quantiles Version 0.1 Date 2015-12-19 Maintainer Could be used to obtain spatial depths, spatial ranks and outliers of multivariate
Package SpatPCA. R topics documented: February 20, Type Package
 Type Package Package SpatPCA February 20, 2018 Title Regularized Principal Component Analysis for Spatial Data Version 1.2.0.0 Date 2018-02-20 URL https://github.com/egpivo/spatpca BugReports https://github.com/egpivo/spatpca/issues
Type Package Package SpatPCA February 20, 2018 Title Regularized Principal Component Analysis for Spatial Data Version 1.2.0.0 Date 2018-02-20 URL https://github.com/egpivo/spatpca BugReports https://github.com/egpivo/spatpca/issues
Package diffeq. February 19, 2015
 Version 1.0-1 Package diffeq February 19, 2015 Title Functions from the book Solving Differential Equations in R Author Karline Soetaert Maintainer Karline Soetaert
Version 1.0-1 Package diffeq February 19, 2015 Title Functions from the book Solving Differential Equations in R Author Karline Soetaert Maintainer Karline Soetaert
Package plw. R topics documented: May 7, Type Package
 Type Package Package plw May 7, 2018 Title Probe level Locally moderated Weighted t-tests. Version 1.40.0 Date 2009-07-22 Author Magnus Astrand Maintainer Magnus Astrand
Type Package Package plw May 7, 2018 Title Probe level Locally moderated Weighted t-tests. Version 1.40.0 Date 2009-07-22 Author Magnus Astrand Maintainer Magnus Astrand
Math 70 Homework 2 Michael Downs. 2(w i b) 0 = (w i b) 0 = 0 = b = 1 n. w i b 2 = nb = = z iz i 2z iaˆd i + ˆd 2 i
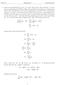 1. I show that minimizing n x i b aĉ i 2 with x i, b, a R n and a 2 = 1 is the same as maximizing the variance of Za. The problem here is finding an n-dimensional best fit line with direction vector a
1. I show that minimizing n x i b aĉ i 2 with x i, b, a R n and a 2 = 1 is the same as maximizing the variance of Za. The problem here is finding an n-dimensional best fit line with direction vector a
Frontier Analysis with R
 Summer School on Mathematical Methods in Finance and Economy June 2010 (slides modified in August 2010) Introduction A first simulated example Analysis of the real data The packages about frontier analysis
Summer School on Mathematical Methods in Finance and Economy June 2010 (slides modified in August 2010) Introduction A first simulated example Analysis of the real data The packages about frontier analysis
Univariate Descriptive Statistics for One Sample
 Department of Psychology and Human Development Vanderbilt University 1 Introduction 2 3 4 5 6 7 8 Introduction Our first step in descriptive statistics is to characterize the data in a single group of
Department of Psychology and Human Development Vanderbilt University 1 Introduction 2 3 4 5 6 7 8 Introduction Our first step in descriptive statistics is to characterize the data in a single group of
Machine Learning - TP
 Machine Learning - TP Nathalie Villa-Vialaneix - nathalie.villa@univ-paris1.fr http://www.nathalievilla.org IUT STID (Carcassonne) & SAMM (Université Paris 1) Formation INRA, Niveau 3 Formation INRA (Niveau
Machine Learning - TP Nathalie Villa-Vialaneix - nathalie.villa@univ-paris1.fr http://www.nathalievilla.org IUT STID (Carcassonne) & SAMM (Université Paris 1) Formation INRA, Niveau 3 Formation INRA (Niveau
A Handbook of Statistical Analyses Using R 2nd Edition. Brian S. Everitt and Torsten Hothorn
 A Handbook of Statistical Analyses Using R 2nd Edition Brian S. Everitt and Torsten Hothorn CHAPTER 7 Logistic Regression and Generalised Linear Models: Blood Screening, Women s Role in Society, Colonic
A Handbook of Statistical Analyses Using R 2nd Edition Brian S. Everitt and Torsten Hothorn CHAPTER 7 Logistic Regression and Generalised Linear Models: Blood Screening, Women s Role in Society, Colonic
A Handbook of Statistical Analyses Using R. Brian S. Everitt and Torsten Hothorn
 A Handbook of Statistical Analyses Using R Brian S. Everitt and Torsten Hothorn CHAPTER 6 Logistic Regression and Generalised Linear Models: Blood Screening, Women s Role in Society, and Colonic Polyps
A Handbook of Statistical Analyses Using R Brian S. Everitt and Torsten Hothorn CHAPTER 6 Logistic Regression and Generalised Linear Models: Blood Screening, Women s Role in Society, and Colonic Polyps
Hypothesis Tests and Confidence Intervals Involving Fitness Landscapes fit by Aster Models By Charles J. Geyer and Ruth G. Shaw Technical Report No.
 Hypothesis Tests and Confidence Intervals Involving Fitness Landscapes fit by Aster Models By Charles J. Geyer and Ruth G. Shaw Technical Report No. 674 School of Statistics University of Minnesota March
Hypothesis Tests and Confidence Intervals Involving Fitness Landscapes fit by Aster Models By Charles J. Geyer and Ruth G. Shaw Technical Report No. 674 School of Statistics University of Minnesota March
coxphmic: An R Package for Sparse Estimation of Cox Proportional Hazards Models via Approximated Information Criteria by Razieh Nabi and Xiaogang Su
 CONTRIBUTED RESEARCH ARTICLE 229 coxphmic: An R Package for Sparse Estimation of Cox Proportional Hazards Models via Approximated Information Criteria by Razieh Nabi and Xiaogang Su Abstract In this paper,
CONTRIBUTED RESEARCH ARTICLE 229 coxphmic: An R Package for Sparse Estimation of Cox Proportional Hazards Models via Approximated Information Criteria by Razieh Nabi and Xiaogang Su Abstract In this paper,
Explore the data. Anja Bråthen Kristoffersen
 Explore the data Anja Bråthen Kristoffersen density 0.2 0.4 0.6 0.8 Probability distributions Can be either discrete or continuous (uniform, bernoulli, normal, etc) Defined by a density function, p(x)
Explore the data Anja Bråthen Kristoffersen density 0.2 0.4 0.6 0.8 Probability distributions Can be either discrete or continuous (uniform, bernoulli, normal, etc) Defined by a density function, p(x)
Logistic Regression. 1 Analysis of the budworm moth data 1. 2 Estimates and confidence intervals for the parameters 2
 Logistic Regression Ulrich Halekoh, Jørgen Vinslov Hansen, Søren Højsgaard Biometry Research Unit Danish Institute of Agricultural Sciences March 31, 2006 Contents 1 Analysis of the budworm moth data 1
Logistic Regression Ulrich Halekoh, Jørgen Vinslov Hansen, Søren Højsgaard Biometry Research Unit Danish Institute of Agricultural Sciences March 31, 2006 Contents 1 Analysis of the budworm moth data 1
Explore the data. Anja Bråthen Kristoffersen Biomedical Research Group
 Explore the data Anja Bråthen Kristoffersen Biomedical Research Group density 0.2 0.4 0.6 0.8 Probability distributions Can be either discrete or continuous (uniform, bernoulli, normal, etc) Defined by
Explore the data Anja Bråthen Kristoffersen Biomedical Research Group density 0.2 0.4 0.6 0.8 Probability distributions Can be either discrete or continuous (uniform, bernoulli, normal, etc) Defined by
The evdbayes Package
 The evdbayes Package April 19, 2006 Version 1.0-5 Date 2006-18-04 Title Bayesian Analysis in Extreme Theory Author Alec Stephenson and Mathieu Ribatet. Maintainer Mathieu Ribatet
The evdbayes Package April 19, 2006 Version 1.0-5 Date 2006-18-04 Title Bayesian Analysis in Extreme Theory Author Alec Stephenson and Mathieu Ribatet. Maintainer Mathieu Ribatet
Finite Mixture Model Diagnostics Using Resampling Methods
 Finite Mixture Model Diagnostics Using Resampling Methods Bettina Grün Johannes Kepler Universität Linz Friedrich Leisch Universität für Bodenkultur Wien Abstract This paper illustrates the implementation
Finite Mixture Model Diagnostics Using Resampling Methods Bettina Grün Johannes Kepler Universität Linz Friedrich Leisch Universität für Bodenkultur Wien Abstract This paper illustrates the implementation
Aster Models and Lande-Arnold Beta By Charles J. Geyer and Ruth G. Shaw Technical Report No. 675 School of Statistics University of Minnesota January
 Aster Models and Lande-Arnold Beta By Charles J. Geyer and Ruth G. Shaw Technical Report No. 675 School of Statistics University of Minnesota January 9, 2010 Abstract Lande and Arnold (1983) proposed an
Aster Models and Lande-Arnold Beta By Charles J. Geyer and Ruth G. Shaw Technical Report No. 675 School of Statistics University of Minnesota January 9, 2010 Abstract Lande and Arnold (1983) proposed an
Item Reliability Analysis
 Item Reliability Analysis Revised: 10/11/2017 Summary... 1 Data Input... 4 Analysis Options... 5 Tables and Graphs... 5 Analysis Summary... 6 Matrix Plot... 8 Alpha Plot... 10 Correlation Matrix... 11
Item Reliability Analysis Revised: 10/11/2017 Summary... 1 Data Input... 4 Analysis Options... 5 Tables and Graphs... 5 Analysis Summary... 6 Matrix Plot... 8 Alpha Plot... 10 Correlation Matrix... 11
Robust Inference in Generalized Linear Models
 Robust Inference in Generalized Linear Models Claudio Agostinelli claudio@unive.it Dipartimento di Statistica Università Ca Foscari di Venezia San Giobbe, Cannaregio 873, Venezia Tel. 041 2347446, Fax.
Robust Inference in Generalized Linear Models Claudio Agostinelli claudio@unive.it Dipartimento di Statistica Università Ca Foscari di Venezia San Giobbe, Cannaregio 873, Venezia Tel. 041 2347446, Fax.
Consider fitting a model using ordinary least squares (OLS) regression:
 Example 1: Mating Success of African Elephants In this study, 41 male African elephants were followed over a period of 8 years. The age of the elephant at the beginning of the study and the number of successful
Example 1: Mating Success of African Elephants In this study, 41 male African elephants were followed over a period of 8 years. The age of the elephant at the beginning of the study and the number of successful
The OmicCircos usages by examples
 The OmicCircos usages by examples Ying Hu and Chunhua Yan October 30, 2017 Contents 1 Introduction 2 2 Input file formats 2 2.1 segment data............................................. 2 2.2 mapping data.............................................
The OmicCircos usages by examples Ying Hu and Chunhua Yan October 30, 2017 Contents 1 Introduction 2 2 Input file formats 2 2.1 segment data............................................. 2 2.2 mapping data.............................................
Hypothesis Tests and Confidence Intervals Involving Fitness Landscapes fit by Aster Models By Charles J. Geyer and Ruth G. Shaw Technical Report No.
 Hypothesis Tests and Confidence Intervals Involving Fitness Landscapes fit by Aster Models By Charles J. Geyer and Ruth G. Shaw Technical Report No. 674 revised School of Statistics University of Minnesota
Hypothesis Tests and Confidence Intervals Involving Fitness Landscapes fit by Aster Models By Charles J. Geyer and Ruth G. Shaw Technical Report No. 674 revised School of Statistics University of Minnesota
Bayesian Regression for a Dirichlet Distributed Response using Stan
 Bayesian Regression for a Dirichlet Distributed Response using Stan Holger Sennhenn-Reulen 1 1 Department of Growth and Yield, Northwest German Forest Research Institute, Göttingen, Germany arxiv:1808.06399v1
Bayesian Regression for a Dirichlet Distributed Response using Stan Holger Sennhenn-Reulen 1 1 Department of Growth and Yield, Northwest German Forest Research Institute, Göttingen, Germany arxiv:1808.06399v1
Prediction problems 3: Validation and Model Checking
 Prediction problems 3: Validation and Model Checking Data Science 101 Team May 17, 2018 Outline Validation Why is it important How should we do it? Model checking Checking whether your model is a good
Prediction problems 3: Validation and Model Checking Data Science 101 Team May 17, 2018 Outline Validation Why is it important How should we do it? Model checking Checking whether your model is a good
f Simulation Example: Simulating p-values of Two Sample Variance Test. Name: Example June 26, 2011 Math Treibergs
 Math 3070 1. Treibergs f Simulation Example: Simulating p-values of Two Sample Variance Test. Name: Example June 26, 2011 The t-test is fairly robust with regard to actual distribution of data. But the
Math 3070 1. Treibergs f Simulation Example: Simulating p-values of Two Sample Variance Test. Name: Example June 26, 2011 The t-test is fairly robust with regard to actual distribution of data. But the
Interpreting Regression
 Interpreting Regression January 10, 2018 Contents Standard error of the estimate Homoscedasticity Using R to make interpretations about regresssion Questions In the tutorial on prediction we used the regression
Interpreting Regression January 10, 2018 Contents Standard error of the estimate Homoscedasticity Using R to make interpretations about regresssion Questions In the tutorial on prediction we used the regression
Holiday Assignment PS 531
 Holiday Assignment PS 531 Prof: Jake Bowers TA: Paul Testa January 27, 2014 Overview Below is a brief assignment for you to complete over the break. It should serve as refresher, covering some of the basic
Holiday Assignment PS 531 Prof: Jake Bowers TA: Paul Testa January 27, 2014 Overview Below is a brief assignment for you to complete over the break. It should serve as refresher, covering some of the basic
Case Study: Modelling Industrial Dryer Temperature Arun K. Tangirala 11/19/2016
 Case Study: Modelling Industrial Dryer Temperature Arun K. Tangirala 11/19/2016 Background This is a case study concerning time-series modelling of the temperature of an industrial dryer. Data set contains
Case Study: Modelling Industrial Dryer Temperature Arun K. Tangirala 11/19/2016 Background This is a case study concerning time-series modelling of the temperature of an industrial dryer. Data set contains
Package sklarsomega. May 24, 2018
 Type Package Package sklarsomega May 24, 2018 Title Measuring Agreement Using Sklar's Omega Coefficient Version 1.0 Date 2018-05-22 Author John Hughes Maintainer John Hughes
Type Package Package sklarsomega May 24, 2018 Title Measuring Agreement Using Sklar's Omega Coefficient Version 1.0 Date 2018-05-22 Author John Hughes Maintainer John Hughes
Counterfactual: An R Package for Counterfactual Analysis by Mingli Chen, Victor Chernozhukov, Iván Fernández-Val and Blaise Melly
 CONTRIBUTED RESEARCH ARTICLE 370 Counterfactual: An R Package for Counterfactual Analysis by Mingli Chen, Victor Chernozhukov, Iván Fernández-Val and Blaise Melly Abstract The Counterfactual package implements
CONTRIBUTED RESEARCH ARTICLE 370 Counterfactual: An R Package for Counterfactual Analysis by Mingli Chen, Victor Chernozhukov, Iván Fernández-Val and Blaise Melly Abstract The Counterfactual package implements
Chapter 5 Exercises 1
 Chapter 5 Exercises 1 Data Analysis & Graphics Using R, 2 nd edn Solutions to Exercises (December 13, 2006) Preliminaries > library(daag) Exercise 2 For each of the data sets elastic1 and elastic2, determine
Chapter 5 Exercises 1 Data Analysis & Graphics Using R, 2 nd edn Solutions to Exercises (December 13, 2006) Preliminaries > library(daag) Exercise 2 For each of the data sets elastic1 and elastic2, determine
Principal Components Analysis using R Francis Huang / November 2, 2016
 Principal Components Analysis using R Francis Huang / huangf@missouri.edu November 2, 2016 Principal components analysis (PCA) is a convenient way to reduce high dimensional data into a smaller number
Principal Components Analysis using R Francis Huang / huangf@missouri.edu November 2, 2016 Principal components analysis (PCA) is a convenient way to reduce high dimensional data into a smaller number
Introduction to R, Part I
 Introduction to R, Part I Basic math, variables, and variable types Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here
Introduction to R, Part I Basic math, variables, and variable types Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here
Package neuralnet. August 16, 2016
 Type Package Title Training of Neural Networks Version 1.33 Date 2016-08-05 Package neuralnet August 16, 2016 Author Stefan Fritsch [aut], Frauke Guenther [aut, cre], Marc Suling [ctb], Sebastian M. Mueller
Type Package Title Training of Neural Networks Version 1.33 Date 2016-08-05 Package neuralnet August 16, 2016 Author Stefan Fritsch [aut], Frauke Guenther [aut, cre], Marc Suling [ctb], Sebastian M. Mueller
Package FDRSeg. September 20, 2017
 Type Package Package FDRSeg September 20, 2017 Title FDR-Control in Multiscale Change-Point Segmentation Version 1.0-3 Date 2017-09-20 Author Housen Li [aut], Hannes Sieling [aut], Timo Aspelmeier [cre]
Type Package Package FDRSeg September 20, 2017 Title FDR-Control in Multiscale Change-Point Segmentation Version 1.0-3 Date 2017-09-20 Author Housen Li [aut], Hannes Sieling [aut], Timo Aspelmeier [cre]
Statistical Equating Methods
 Statistical Equating Methods Anthony Albano January 7, 2011 Abstract The R package equate (Albano, 2011) contains functions for non-irt equating under random groups and nonequivalent groups designs. This
Statistical Equating Methods Anthony Albano January 7, 2011 Abstract The R package equate (Albano, 2011) contains functions for non-irt equating under random groups and nonequivalent groups designs. This
Solving partial differential equations, using R package ReacTran
 Solving partial differential equations, using R package ReacTran Karline Soetaert and Filip Meysman Centre for Estuarine and Marine Ecology Netherlands Institute of Ecology The Netherlands Abstract R -package
Solving partial differential equations, using R package ReacTran Karline Soetaert and Filip Meysman Centre for Estuarine and Marine Ecology Netherlands Institute of Ecology The Netherlands Abstract R -package
Bayesian inference for a population growth model of the chytrid fungus Philipp H Boersch-Supan, Sadie J Ryan, and Leah R Johnson September 2016
 Bayesian inference for a population growth model of the chytrid fungus Philipp H Boersch-Supan, Sadie J Ryan, and Leah R Johnson September 2016 1 Preliminaries This vignette illustrates the steps needed
Bayesian inference for a population growth model of the chytrid fungus Philipp H Boersch-Supan, Sadie J Ryan, and Leah R Johnson September 2016 1 Preliminaries This vignette illustrates the steps needed
Univariate Analysis of Variance
 Univariate Analysis of Variance Output Created Comments Input Missing Value Handling Syntax Resources Notes Data Active Dataset Filter Weight Split File N of Rows in Working Data File Definition of Missing
Univariate Analysis of Variance Output Created Comments Input Missing Value Handling Syntax Resources Notes Data Active Dataset Filter Weight Split File N of Rows in Working Data File Definition of Missing
G.M. Berberich, C.F. Dormann, D. Klimetzek, M.B. Berberich, N.J. Sanders & A.M. Ellison. Appendix S2
 G.M. Berberich, C.F. Dormann, D. Klimetzek, M.B. Berberich, N.J. Sanders & A.M. Ellison Appendix S2 The supplement material includes the data set, the commented R-code for all our analyses and figures
G.M. Berberich, C.F. Dormann, D. Klimetzek, M.B. Berberich, N.J. Sanders & A.M. Ellison Appendix S2 The supplement material includes the data set, the commented R-code for all our analyses and figures
Package clogitboost. R topics documented: December 21, 2015
 Package December 21, 2015 Type Package Title Boosting Conditional Logit Model Version 1.1 Date 2015-12-09 Author Haolun Shi and Guosheng Yin Maintainer A set of functions to fit a boosting conditional
Package December 21, 2015 Type Package Title Boosting Conditional Logit Model Version 1.1 Date 2015-12-09 Author Haolun Shi and Guosheng Yin Maintainer A set of functions to fit a boosting conditional
Leslie matrices and Markov chains.
 Leslie matrices and Markov chains. Example. Suppose a certain species of insect can be divided into 2 classes, eggs and adults. 10% of eggs survive for 1 week to become adults, each adult yields an average
Leslie matrices and Markov chains. Example. Suppose a certain species of insect can be divided into 2 classes, eggs and adults. 10% of eggs survive for 1 week to become adults, each adult yields an average
A Handbook of Statistical Analyses Using R 2nd Edition. Brian S. Everitt and Torsten Hothorn
 A Handbook of Statistical Analyses Using R 2nd Edition Brian S. Everitt and Torsten Hothorn CHAPTER 12 Analysing Longitudinal Data I: Computerised Delivery of Cognitive Behavioural Therapy Beat the Blues
A Handbook of Statistical Analyses Using R 2nd Edition Brian S. Everitt and Torsten Hothorn CHAPTER 12 Analysing Longitudinal Data I: Computerised Delivery of Cognitive Behavioural Therapy Beat the Blues
A Handbook of Statistical Analyses Using R 3rd Edition. Torsten Hothorn and Brian S. Everitt
 A Handbook of Statistical Analyses Using R 3rd Edition Torsten Hothorn and Brian S. Everitt CHAPTER 12 Quantile Regression: Head Circumference for Age 12.1 Introduction 12.2 Quantile Regression 12.3 Analysis
A Handbook of Statistical Analyses Using R 3rd Edition Torsten Hothorn and Brian S. Everitt CHAPTER 12 Quantile Regression: Head Circumference for Age 12.1 Introduction 12.2 Quantile Regression 12.3 Analysis
BIOL 458 BIOMETRY Lab 9 - Correlation and Bivariate Regression
 BIOL 458 BIOMETRY Lab 9 - Correlation and Bivariate Regression Introduction to Correlation and Regression The procedures discussed in the previous ANOVA labs are most useful in cases where we are interested
BIOL 458 BIOMETRY Lab 9 - Correlation and Bivariate Regression Introduction to Correlation and Regression The procedures discussed in the previous ANOVA labs are most useful in cases where we are interested
Package bpp. December 13, 2016
 Type Package Package bpp December 13, 2016 Title Computations Around Bayesian Predictive Power Version 1.0.0 Date 2016-12-13 Author Kaspar Rufibach, Paul Jordan, Markus Abt Maintainer Kaspar Rufibach Depends
Type Package Package bpp December 13, 2016 Title Computations Around Bayesian Predictive Power Version 1.0.0 Date 2016-12-13 Author Kaspar Rufibach, Paul Jordan, Markus Abt Maintainer Kaspar Rufibach Depends
Getting to the Roots of Quadratics
 NAME BACKGROUND Graphically: The real roots of a function are the x-coordinates of the points at which the graph of the function intercepts/crosses the x-axis. For a quadratic function, whose graph is
NAME BACKGROUND Graphically: The real roots of a function are the x-coordinates of the points at which the graph of the function intercepts/crosses the x-axis. For a quadratic function, whose graph is
A Handbook of Statistical Analyses Using R 2nd Edition. Brian S. Everitt and Torsten Hothorn
 A Handbook of Statistical Analyses Using R 2nd Edition Brian S. Everitt and Torsten Hothorn CHAPTER 12 Analysing Longitudinal Data I: Computerised Delivery of Cognitive Behavioural Therapy Beat the Blues
A Handbook of Statistical Analyses Using R 2nd Edition Brian S. Everitt and Torsten Hothorn CHAPTER 12 Analysing Longitudinal Data I: Computerised Delivery of Cognitive Behavioural Therapy Beat the Blues
GENERALIZED ERROR DISTRIBUTION
 CHAPTER 21 GENERALIZED ERROR DISTRIBUTION 21.1 ASSIGNMENT Write R functions for the Generalized Error Distribution, GED. Nelson [1991] introduced the Generalized Error Distribution for modeling GARCH time
CHAPTER 21 GENERALIZED ERROR DISTRIBUTION 21.1 ASSIGNMENT Write R functions for the Generalized Error Distribution, GED. Nelson [1991] introduced the Generalized Error Distribution for modeling GARCH time
EDAMI DIMENSION REDUCTION BY PRINCIPAL COMPONENT ANALYSIS
 EDAMI DIMENSION REDUCTION BY PRINCIPAL COMPONENT ANALYSIS Mario Romanazzi October 29, 2017 1 Introduction An important task in multidimensional data analysis is reduction in complexity. Recalling that
EDAMI DIMENSION REDUCTION BY PRINCIPAL COMPONENT ANALYSIS Mario Romanazzi October 29, 2017 1 Introduction An important task in multidimensional data analysis is reduction in complexity. Recalling that
Chapter 8 Conclusion
 1 Chapter 8 Conclusion Three questions about test scores (score) and student-teacher ratio (str): a) After controlling for differences in economic characteristics of different districts, does the effect
1 Chapter 8 Conclusion Three questions about test scores (score) and student-teacher ratio (str): a) After controlling for differences in economic characteristics of different districts, does the effect
A Brief Introduction To. GRTensor. On MAPLE Platform. A write-up for the presentation delivered on the same topic as a part of the course PHYS 601
 A Brief Introduction To GRTensor On MAPLE Platform A write-up for the presentation delivered on the same topic as a part of the course PHYS 601 March 2012 BY: ARSHDEEP SINGH BHATIA arshdeepsb@gmail.com
A Brief Introduction To GRTensor On MAPLE Platform A write-up for the presentation delivered on the same topic as a part of the course PHYS 601 March 2012 BY: ARSHDEEP SINGH BHATIA arshdeepsb@gmail.com
A Handbook of Statistical Analyses Using R 3rd Edition. Torsten Hothorn and Brian S. Everitt
 A Handbook of Statistical Analyses Using R 3rd Edition Torsten Hothorn and Brian S. Everitt CHAPTER 10 Scatterplot Smoothers and Generalized Additive Models: The Men s Olympic 1500m, Air Pollution in
A Handbook of Statistical Analyses Using R 3rd Edition Torsten Hothorn and Brian S. Everitt CHAPTER 10 Scatterplot Smoothers and Generalized Additive Models: The Men s Olympic 1500m, Air Pollution in
Package pearson7. June 22, 2016
 Version 1.0-2 Date 2016-06-21 Package pearson7 June 22, 2016 Title Maximum Likelihood Inference for the Pearson VII Distribution with Shape Parameter 3/2 Author John Hughes Maintainer John Hughes
Version 1.0-2 Date 2016-06-21 Package pearson7 June 22, 2016 Title Maximum Likelihood Inference for the Pearson VII Distribution with Shape Parameter 3/2 Author John Hughes Maintainer John Hughes
COUNTERFACTUAL ANALYSIS IN R: A VIGNETTE
 COUNTERFACTUAL ANALYSIS IN R: A VIGNETTE MINGLI CHEN, VICTOR CHERNOZHUKOV, IVÁN FERNÁNDEZ-VAL, AND BLAISE MELLY Abstract. The R package Counterfactual implements the estimation and inference methods of
COUNTERFACTUAL ANALYSIS IN R: A VIGNETTE MINGLI CHEN, VICTOR CHERNOZHUKOV, IVÁN FERNÁNDEZ-VAL, AND BLAISE MELLY Abstract. The R package Counterfactual implements the estimation and inference methods of
Interpreting Regression
 Interpreting Regression January 27, 2019 Contents Standard error of the estimate Homoscedasticity Using R to make interpretations about regresssion Questions In the tutorial on prediction we used the regression
Interpreting Regression January 27, 2019 Contents Standard error of the estimate Homoscedasticity Using R to make interpretations about regresssion Questions In the tutorial on prediction we used the regression
Class 04 - Statistical Inference
 Class 4 - Statistical Inference Question 1: 1. What parameters control the shape of the normal distribution? Make some histograms of different normal distributions, in each, alter the parameter values
Class 4 - Statistical Inference Question 1: 1. What parameters control the shape of the normal distribution? Make some histograms of different normal distributions, in each, alter the parameter values
cor(dataset$measurement1, dataset$measurement2, method= pearson ) cor.test(datavector1, datavector2, method= pearson )
 Tutorial 7: Correlation and Regression Correlation Used to test whether two variables are linearly associated. A correlation coefficient (r) indicates the strength and direction of the association. A correlation
Tutorial 7: Correlation and Regression Correlation Used to test whether two variables are linearly associated. A correlation coefficient (r) indicates the strength and direction of the association. A correlation
GPoM : 2 PreProcessing
 GPoM : 2 PreProcessing Sylvain Mangiarotti & Mireille Huc 2018-07-26 Pre-processing for global modelling To obtain Ordinary Differential Equations (ODEs) from time series requires the computation of its
GPoM : 2 PreProcessing Sylvain Mangiarotti & Mireille Huc 2018-07-26 Pre-processing for global modelling To obtain Ordinary Differential Equations (ODEs) from time series requires the computation of its
Metric Predicted Variable on One Group
 Metric Predicted Variable on One Group Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Prior Homework
Metric Predicted Variable on One Group Tim Frasier Copyright Tim Frasier This work is licensed under the Creative Commons Attribution 4.0 International license. Click here for more information. Prior Homework
Contents. Introduction 2. PART A - data import & R session setup 3 Step 1 - read in the data load the libraries sessioninfo...
 R code for The human immune system is robustly maintained in multiple stable equilibriums shaped by age and cohabitation Ed Carr, on behalf of co-authors 10 September 2015 Contents Introduction 2 PART
R code for The human immune system is robustly maintained in multiple stable equilibriums shaped by age and cohabitation Ed Carr, on behalf of co-authors 10 September 2015 Contents Introduction 2 PART
ensemblebma: An R Package for Probabilistic Forecasting using Ensembles and Bayesian Model Averaging
 ensemblebma: An R Package for Probabilistic Forecasting using Ensembles and Bayesian Model Averaging Chris Fraley, Adrian E. Raftery Tilmann Gneiting, J. McLean Sloughter Technical Report No. 516R Department
ensemblebma: An R Package for Probabilistic Forecasting using Ensembles and Bayesian Model Averaging Chris Fraley, Adrian E. Raftery Tilmann Gneiting, J. McLean Sloughter Technical Report No. 516R Department
Logistic Regression. 0.1 Frogs Dataset
 Logistic Regression We move now to the classification problem from the regression problem and study the technique ot logistic regression. The setting for the classification problem is the same as that
Logistic Regression We move now to the classification problem from the regression problem and study the technique ot logistic regression. The setting for the classification problem is the same as that
Inferences on Linear Combinations of Coefficients
 Inferences on Linear Combinations of Coefficients Note on required packages: The following code required the package multcomp to test hypotheses on linear combinations of regression coefficients. If you
Inferences on Linear Combinations of Coefficients Note on required packages: The following code required the package multcomp to test hypotheses on linear combinations of regression coefficients. If you
ftsa: An R Package for Analyzing Functional Time Series by Han Lin Shang
 CONTRIBUTED RESEARCH ARTICLES 64 ftsa: An R Package for Analyzing Functional Time Series by Han Lin Shang Abstract Recent advances in computer recording and storing technology have tremendously increased
CONTRIBUTED RESEARCH ARTICLES 64 ftsa: An R Package for Analyzing Functional Time Series by Han Lin Shang Abstract Recent advances in computer recording and storing technology have tremendously increased
COUNTERFACTUAL ANALYSIS IN R: A VIGNETTE
 COUNTERFACTUAL ANALYSIS IN R: A VIGNETTE MINGLI CHEN, VICTOR CHERNOZHUKOV, IVÁN FERNÁNDEZ-VAL, AND BLAISE MELLY Abstract. The R package Counterfactual implements the estimation and inference methods of
COUNTERFACTUAL ANALYSIS IN R: A VIGNETTE MINGLI CHEN, VICTOR CHERNOZHUKOV, IVÁN FERNÁNDEZ-VAL, AND BLAISE MELLY Abstract. The R package Counterfactual implements the estimation and inference methods of
arxiv: v1 [stat.co] 7 Jul 2017
![arxiv: v1 [stat.co] 7 Jul 2017 arxiv: v1 [stat.co] 7 Jul 2017](/thumbs/75/71512890.jpg) Object-Oriented Software for Functional Data Clara Happ Department of Statistics, LMU Munich, Munich, Germany. arxiv:77.9v [stat.co] 7 Jul 7 Abstract This paper introduces the fundata R package as an object-oriented
Object-Oriented Software for Functional Data Clara Happ Department of Statistics, LMU Munich, Munich, Germany. arxiv:77.9v [stat.co] 7 Jul 7 Abstract This paper introduces the fundata R package as an object-oriented

 Homework 6 Solutions set.seed(1) library(mvtnorm) samp.theta
Homework 6 Solutions set.seed(1) library(mvtnorm) samp.theta