InChI/InChIKey vs. NCI/CADD Structure Identifiers: A comparison
|
|
|
- Rosemary Blair
- 5 years ago
- Views:
Transcription
1 InChI/InChIKey vs. CI/CADD Structure Identifiers: A comparison Markus Sitzmann Computer-Aided Drug Design Group (CI/CADD), Laboratory of Medicinal Chemistry, CI-Frederick, I, DS Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
2 The Adaption and Use of the IUPAC InChI/InChIKey Chemical Structure Lookup Service 74 million structure records 46 million unique structures InChI/InChIKey Std. InChI/InChIKey CI/CADD Identifiers FICTS FICuS uuuuu Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
3 Unique Representation of Chemical Structures CI/CADD Structure Identifiers based on hashcodes calculated by the chemoinformatics toolkit CACTVS 2 CACTVS hashcodes: 9850FD9F9E2B4E25 represent a chemical structure uniquely as 16-digit hexadecimal number (64-bit unsigned) have a high sensitivity to structural features of a compound change if connectivity changes Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
4 2 6C16DE2351F9FF50 tautomers 2 E92E4BA2869F3611 stereoisomers 2 8A7AD1EB498CC76A 2 salt - 2 charged form a ECEF579D7DF025A 9850FD9F9E2B4E25 A3DAE DDE4 a 2 8F7A1DE5A733F0E0 errors isotope 15 2 B2FDA68AEDA06DB E1AF41497B6 Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
5 Unique Representation of Chemical Structures CI/CADD Structure Identifiers MDL Molfile MDL SDF SMILES ChemDraw cdx PDB input structure structure normalization parent structure hashcode calculation E_ASISY CI/CADD Identifier MDL SDF SMILES database Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
6 CI/CADD Structure Identifiers Structure ormalization adjustable levels of sensitivity: Fragments Isotopes Charges Tautomers Stereochemistry sensitive sensitive sensitive sensitive sensitive - a + D D D D D D C 2 C 2 keep only largest organic fragment ignore isotope labels uncharge find canonical tautomer discard stereo information 2 C 2 un-sensitive un-sensitive un-sensitive un-sensitive un-sensitive Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
7 CI/CADD Structure Identifiers Structure ormalization Fragments Isotopes Charges sensitive sensitive sensitive - a + D D D D D D Tautomers sensitive Stereochemistry sensitive C 2 C 2 2 C 2 un-sensitive un-sensitive un-sensitive un-sensitive Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
8 CI/CADD Structure Identifiers Structure ormalization FICTS identifier: representation of the exact drawing Fragments Isotopes Charges sensitive sensitive sensitive - a + D D D D D D F I C Tautomers sensitive = T = Stereochemistry sensitive C 2 S C 2 2 C 2 un-sensitive un-sensitive un-sensitive un-sensitive un-sensitive Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
9 CI/CADD Structure Identifiers Structure ormalization FICuS identifier: comes closest to how a chemist perceives a compound Fragments Isotopes Charges sensitive sensitive sensitive - a + D D D D D D Tautomers sensitive = Stereochemistry sensitive C 2 C 2 F I C = u S 2 C 2 un-sensitive un-sensitive un-sensitive un-sensitive un-sensitive Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
10 CI/CADD Structure Identifier Structure ormalization uuuuu identifier: closely related forms of the same compound Fragments Isotopes Charges Tautomers Stereochemistry sensitive sensitive sensitive sensitive sensitive - a + D D D D D D = C = 2 C 2 = u = = = = = u u u u 2 C 2 un-sensitive un-sensitive un-sensitive un-sensitive un-sensitive Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
11 CI/CADD Structure Identifier Structure ormalization normalize or discard stereo information define canonical tautomer n FICTS define canonical resonance form/ protonation state n d FICTu FICuS input structure d n FICuu uuuts parent structures correct structure: add hydrogen atoms correct functional groups correct metal atom bonds n d uuutu uuuus get largest fragment & uncharge: delete complex center get largest organic fragment delete radical center uncharge structure d discard isotope labels uuuuu Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
12 CI/CADD Structure Identifier FD9F9E2B4E25-FICTS FD9F9E2B4E25-FICuS FD9F9E2B4E25-uuuuu <CACTVS hashcode (E_ASISY)>-<tag>-<version>-<checksum> Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
13 2 6C16DE2351F9FF50-FICTS tautomers 2 E92E4BA2869F3611-FICTS stereoisomers 2 8A7AD1EB498CC76A-FICTS 2 salt - a charged form E5F83F10C5DB080A-FICTS FD9F9E2B4E25-FICTS FICTS errors a A3DAE DDE4-FICTS isotope 15 2 E5F83F10C5DB080A-FICTS B2FDA68AEDA06DB9-FICTS 9850FD9F9E2B4E25-FICTS Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
14 2 9850FD9F9E2B4E25-FICuS tautomers 2 E92E4BA2869F3611-FICuS stereoisomers 2 8A7AD1EB498CC76A-FICuS 2 salt - a charged form E5F83F10C5DB080A-FICuS FD9F9E2B4E25-FICuS FICuS errors a A3DAE DDE4-FICuS isotope 15 2 E5F83F10C5DB080A-FICuS B2FDA68AEDA06DB9-FICuS 9850FD9F9E2B4E25-FICuS Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
15 2 9850FD9F9E2B4E25-uuuuu tautomers FD9F9E2B4E25-uuuuu stereoisomers FD9F9E2B4E25-uuuuu 2 salt - a charged form 9850FD9F9E2B4E25-uuuuu 9850FD9F9E2B4E25-uuuuu 9850FD9F9E2B4E25-uuuuu 2 uuuuu errors a isotope FD9F9E2B4E25-uuuuu 9850FD9F9E2B4E25-uuuuu 9850FD9F9E2B4E25-FICuS Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
16 2 DVDQJCIGZP-UFFFAYSA- tautomers 2 DVDQJCIGZP-RXMQYKEDSA- stereoisomers 2 DVDQJCIGZP-YFKPBYRVSA- 2 salt - a charged form UPKBYGGMJTIM-UFFFAYSA-M DVDQJCIGZP-UFFFAYSA- DVDQJCIGZP-UFFFAYSA- 2 Std. InChIKey errors a isotope UPKBYGGMJTIM-UFFFAYSA-M DVDQJCIGZP-UFFFAYSA- DVDQJCIGZP-CDYZYAPPSA- Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
17 Structure ormalization Tautomers canonical tautomer? Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
18 Structure ormalization Tautomers CACTVS: generation of all formal tautomers for a given organic compound (prototropic tautomerism) rule set of 21 transforms encoded as (CACTVS-extended) SMIRKS types of tautomerism covered: 1.3, 1.5 keto/enol imine/enamine imine/amine lactam/lactim 1.3, 1.5, 1.7, 1.11 hydrogen atom shift on (aromatic) heteroatoms keten/ynol nitro/aci-nitro nitroso/oxime special cases: cyanic/iso-cyanic acid, phosphonic acid, formamidinesulfonic acid, isocyanide, furanones and more Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
19 Structure ormalization Tautomers 21 SMIRKS transforms, examples: transform: 1.3 keto-enol [,S,Se,Te;X1:1]=[Cx1:2][CX4R{0-2}:3][#1:4]>> [#1:4][,S,Se,Te;X2:1][Cx1,cx1:2]=[C,cx1,cx0:3] transform: 1.3 heteroatom shift [,n,s,s,,o,se,te:1]=[x2,nx2,c,c,p,p:2] [,n,s,,se,te:3][#1:4]>>[#1:4][,n,s,,se,te:1] [X2,nX2,C,c,P,p:2]=[,n,S,s,,o,Se,Te:3] transform: 1.5 heteroatom shift [nx2,x2,s,,se,te:1]=[c,c,nx2,x2:6][c,c:5]=[c,c,nx2:2] [,n,s,s,,o,se,te:3][#1:4]>>[#1:4][,n,s,,se,te:1] [C,c,nX2,X2:6]=[C,c:5][C,c,nX2:2]=[X2,S,,Se,Te:3] Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
20 Structure ormalization Tautomers guanine A6199E68A788F2F5-FICTS 67196F0B20B1D934-FICTS D979CF9770AC0BA5-FICTS 675R4FCC50F45026-FICTS 959B273B619C709F-FICTS AD375920BE60DAD-FICTS BCCDA7D0CDACF120-FICTS CE8F480C11DBFC4F-FICTS 61248C4A7D045A47-FICTS 0B345B47F FICTS 181CA9BCE3EF47F4-FICTS D46A1E6500B06AB6-FICTS 56FFE8B5619FB01-FICTS F802E527EC5C61BF-FICTS EF060DA9D97091DE-FICTS UYTPUPDQBUYGX-UFFFAYSA- BCCDA7D0CDACF120-FICuS Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
21 Structure ormalization Tautomerism & Stereochemistry methyl propenyl ketone E Z Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
22 Structure ormalization Tautomerism & Stereochemistry methyl propenyl ketone E tautomer tautomer Z Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
23 Structure ormalization Tautomerism & Stereochemistry methyl propenyl ketone E tautomer 76D03F08ACDF6C0C-FICuS FICUS disregards stereochemistry on double bonds if the double bond is not located during tautomer generation. Z tautomer Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
24 InChI/InChIKey - CI/CADD Identifier comparison Tautomerism & Stereochemistry InChI=1S/C58/c (2)6/h3-4,1-23 LABTWGUMFABVFG-UFFFAYSA- E methyl propenyl ketone InChI=1S/C58/c (2)6/h3-4,1-23/b4-3+ LABTWGUMFABVFG-EGZZKSA- tautomer 76D03F08ACDF6C0C-FICuS InChI=1S/C58/c (2)6/h3-4,6,12,23/b5-4- LYGWZVQSCPYDG-PLGDYQASA- FICUS disregards stereochemistry on double bonds if the double bond is not located during tautomer generation. Z tautomer InChI=1S/C58/c (2)6/h3-4,1-23/b4-3- LABTWGUMFABVFG-ARJAWSKDSA- Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
25 InChI/InChIKey - CI/CADD Identifier comparison Tautomerism & Stereochemistry methyl propenyl ketone E 821D8C17ACE5040E-FICTS tautomer 76D03F08ACDF6C0C-FICTS 6EB4AA2BAA11965F-FICTS FICTS sees four different structures tautomer Z FICTS Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
26 Structure ormalization Charges in Resonance Systems uncharge F3A27F03AE77A722 62FADCB01F197FC9 canonical resonance structure? problem! different protonation states uncharge F3A27F03AE77A722 2E011EE4519F7920 Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
27 Structure ormalization Charges in Resonance Systems generation of all formal resonance structures for a given (charged) organic compound rule set of 14 transforms encoded as (CACTVS-extended) SMIRKS shifting of charges: 5 rules recombination of charges: 5 rules separation of charges: 4 rules Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
28 Structure ormalization Charges in Resonance Systems münchnones: 1.2 recombination separation (pentavalent atom) 1.3 shift 1.2 recombination 1.3 shift 1.2 shift 1.3 recombination 1.3 shift 1.3 shift 1.3 shift 1.3 shift (no plausible unpolarized resonance structure can be drawn) IUYUGWCTLFFCL-UFFFAYSA- F68AC07DE0D3379F-FICuS Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
29 InChI/InChIKey - CI/CADD Identifier comparison»chemical Structure Lookup Service«Database PubChem database (including pen CI database, EPA DSSTox databases, IAID IV databases, IST Webbook, LM ChemIDplus, ChemSpider ) Chemavigator iresearch Library (compilation of commercially available screening compounds from ~250 international chemistry suppliers) thers ~10% Chemav. iresearch Lib. ~43% PubChem ~47% Commercial Sources / thers (Asinex, Comgenex, ) 74 million structure records (~46 million unique structures) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
30 InChI/InChIKey - CI/CADD Identifier comparison Unique Structure Counts structure records registered in CSLS: 74.2 million successful calculation of: Standard InChI/InChIKey: 73.8 million records CI/CADD Structure Identifiers: 73.7 million records unique structure counts (compound sets) Standard InChI/InChIKey: FICTS Identifier FICuS Identifier Standard InChIKey (first block) uuuuu Identifier 48,027,940 48,023,835 46,715,521 43,055,589 41,671,010 Standard InChI/InChIKeys were calculated by stdinchi-1 (Linux i-386 executable) from the original SD file records Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
31 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison FICuS compound set (46.7 million unique) Standard InChI/InChIKey set calculated by stdinchi-1 (73.8 million, 48.0 million unique) original structure record set (74.2 million) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
32 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 1 conflicts? FICuS compound set (46.7 million unique) Standard InChI/InChIKey set calculated by stdinchi-1 (73.8 million, 48.0 million unique) original structure record set (74.2 million) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
33 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 1 conflicts? FICuS compound set (46.7 million unique) Standard InChI/InChIKey set calculated by stdinchi-1 (73.8 million, 48.0 million unique) original structure record set (74.2 million) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
34 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 2 same InChI/InChIKey? Standard InChI/InChIKey calculated by CACTVS from FICuS compound structure FICuS compound set (46.7 million unique) Standard InChI/InChIKey set calculated by stdinchi-1 (73.8 million, 48.0 million unique) original structure record set (74.2 million) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
35 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 1 no conflicts between Std. InChI/InChIKey and FICuS structure records (million records) all structure records 73.7 FICuS linked to a single InChI/InChIKey 62.3 (84.5%) both linked to a single structure record 34.4 (46.9%) both linked to multiple structure records 27.9 (38.0%) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
36 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 1 conflicts between Std. InChI/InChIKey and FICuS structure records (million records) all structure records 73.7 FICuS is linked to multiple InChI/InChIKeys or vice versa 10.9 (14.7%) one FICuS is linked to multiple InChI/InChIKeys 6.8 (9.2%) one InChI/InChIKey is linked to multiple FICuS 4.1 (5.5%) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
37 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 1 conflicts between Std. InChI/InChIKey and FICuS structure records (million records) all structure records 73.7 FICuS is linked to multiple InChI/InChIKeys or vice versa 10.9 (14.7%) one FICuS is linked to multiple InChI/InChIKeys number of InChIKey first block one InChI/InChIKey is linked to multiple FICuS 4.1 number of InChIKey first block 1.0 (9.2%) (3.1%) (5.5%) (1.3%) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
38 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 2 same InChI/InChIKey? compounds (unique structures) (million records) structure records (million records) all compounds InChI changes all records InChI changes FICTS (7.9%) 4.6 (6.2%) FICuS (13.7%) (12.7%) uuuuu (28.6%) 21.9 (29.7%) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
39 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison 2 same InChI/InChIKey? compounds (unique structures) (million records) structure records (million records) all compounds InChI changes all records InChI changes FICTS (7.9%) 4.6 (6.2%) FICuS (13.7%) (12.7%) uuuuu 41.6 vs. InChIKey first block 11.9 (28.6%) 21.9 (29.7%) 3.2 (7.6%) 6.3 (8.4%) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
40 InChI/InChIKey - CI/CADD Identifier comparison Detailed Comparison compound classification (formal) tautomer count > 1 (formal) tautomer count > 3 (formal) tautomer count > 10 full stereo contains metal atoms metal complexes salt has resonance charges inorganic occurrence in FICuS set 56.4% 25.4% 5.5% 25.7% 0.8% 0.2% 1.0% 0.2% 0.1% occurrence in FICuS subset (InChI changes) 14.5% 18.5% 28.9% 16.9% 34.5% 52.1% 18.6% 52.1% 33.9% Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
41 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ FICuS: 12 different structure records linked to this structure Std. InChI/InChIKey (stdinchi-1): calculates 3 different strings/keys for these 12 structure records (all have the same connectivity layer/first block) all of these 3 StdInChI/InChIKey differ from the StdInChI/InChIKey calculated after FICuS normalization (including connectivity layer/ first block) Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
42 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ E Z Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
43 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ tautomer: E Z Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
44 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ tautomer: tautomeric interconversion? E Z Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
45 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ R tautomeric interconversion? S tautomer: tautomeric interconversion? E Z Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
46 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ R tautomeric interconversion? S tautomer: tautomeric interconversion? E Z Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
47 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ ow many structures? tautomer: R ZIC tautomeric interconversion? S tautomeric interconversion? E ChemBlock A3422/ Chemavigator IST MS-Lib Z Chemavigator Comparison Chemavigator Standard InChI/InChIKeys CI/CADD Structure Identifiers
48 InChI/InChIKey - CI/CADD Identifier comparison ChemBlock A3422/ ow many structures? R tautomeric interconversion? S tautomer: FICuS parent structure tautomeric interconversion? E InChIKey A Z InChIKey C same connectivity layer/block InChIKey B Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
49 The Adaption and Use of the IUPAC InChI/InChIKey Chemical Structure Lookup Service 74 million structure records 46 million unique structures InChI/InChIKey Std. InChI/InChIKey CI/CADD Identifiers FICTS FICuS uuuuu Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
50 Web Service Chemical Structure REST Service (beta) URL scheme: returns plain text/gif image if the structure identifier is not resolvable: http 404 status code Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
51 Acknowledgments CADD Group, LMC, CI Marc icklaus Igor V. Filippov Chemavigator Scott utton Tad urst CACTVS, Xemistry Gmb Wolf-Dietrich Ihlenfeldt Thanks to all database providers Thanks to the InChI Team ur web site: Comparison Standard InChI/InChIKeys - CI/CADD Structure Identifiers
The NCI/CADD Group's InChI Usage and Analysis of Tautomerism for InChI V2
 The NCI/CADD Group's InChI Usage and Analysis of Tautomerism for InChI V2 Marc C. Nicklaus Computer-Aided Drug Design (CADD) Group Chemical Biology Laboratory Center for Cancer Research National Cancer
The NCI/CADD Group's InChI Usage and Analysis of Tautomerism for InChI V2 Marc C. Nicklaus Computer-Aided Drug Design (CADD) Group Chemical Biology Laboratory Center for Cancer Research National Cancer
The IBM Patent Data Donation to NIH, and its Integration in the NCI/CADD Database and Web Services
 The IBM Patent Data Donation to NIH, and its Integration in the NCI/CADD Database and Web Services Marc C. Nicklaus Computer-Aided Drug Design Group, Chemical Biology Laboratory, Frederick National Laboratory
The IBM Patent Data Donation to NIH, and its Integration in the NCI/CADD Database and Web Services Marc C. Nicklaus Computer-Aided Drug Design Group, Chemical Biology Laboratory, Frederick National Laboratory
Tautomerism in chemical information management systems
 Tautomerism in chemical information management systems Dr. Wendy A. Warr http://www.warr.com Tautomerism in chemical information management systems Author: Wendy A. Warr DOI: 10.1007/s10822-010-9338-4
Tautomerism in chemical information management systems Dr. Wendy A. Warr http://www.warr.com Tautomerism in chemical information management systems Author: Wendy A. Warr DOI: 10.1007/s10822-010-9338-4
Canonical Line Notations
 Canonical Line otations InChI vs SMILES Krisztina Boda verview Compound naming InChI SMILES Molecular equivalency Isomorphism Kekule Tautomers Finding duplicates What s Your ame? 1. Unique numbers CAS
Canonical Line otations InChI vs SMILES Krisztina Boda verview Compound naming InChI SMILES Molecular equivalency Isomorphism Kekule Tautomers Finding duplicates What s Your ame? 1. Unique numbers CAS
The IUPAC Chemical Identifier
 The IUPAC Chemical Identifier Steve Stein, Steve eller, Dmitrii Tchekhovskoi ational Institute of Standards and Technology Gaithersburg, MD, USA CAS/IUPAC Conference on Chemical Identifiers and XML for
The IUPAC Chemical Identifier Steve Stein, Steve eller, Dmitrii Tchekhovskoi ational Institute of Standards and Technology Gaithersburg, MD, USA CAS/IUPAC Conference on Chemical Identifiers and XML for
InChI keys as standard global identifiers in chemistry web services. Russ Hillard ACS, Salt Lake City March 2009
 InChI keys as standard global identifiers in chemistry web services Russ Hillard ACS, Salt Lake City March 2009 Context of this talk We have created a web service That aggregates sources built independently
InChI keys as standard global identifiers in chemistry web services Russ Hillard ACS, Salt Lake City March 2009 Context of this talk We have created a web service That aggregates sources built independently
DECEMBER 2014 REAXYS R201 ADVANCED STRUCTURE SEARCHING
 DECEMBER 2014 REAXYS R201 ADVANCED STRUCTURE SEARCHING 1 NOTES ON REAXYS R201 THIS PRESENTATION COMMENTS AND SUMMARY Outlines how to: a. Perform Substructure and Similarity searches b. Use the functions
DECEMBER 2014 REAXYS R201 ADVANCED STRUCTURE SEARCHING 1 NOTES ON REAXYS R201 THIS PRESENTATION COMMENTS AND SUMMARY Outlines how to: a. Perform Substructure and Similarity searches b. Use the functions
Introduction to Chemoinformatics and Drug Discovery
 Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
Organometallics & InChI. August 2017
 Organometallics & InChI August 2017 The Cambridge Structural Database 900,000+ small-molecule crystal structures Over 60,000 datasets deposited annually Enriched and annotated by experts Structures available
Organometallics & InChI August 2017 The Cambridge Structural Database 900,000+ small-molecule crystal structures Over 60,000 datasets deposited annually Enriched and annotated by experts Structures available
Capturing Chemistry. What you see is what you get In the world of mechanism and chemical transformations
 Capturing Chemistry What you see is what you get In the world of mechanism and chemical transformations Dr. Stephan Schürer ead of Intl. Sci. Content Libraria, Inc. sschurer@libraria.com Distribution of
Capturing Chemistry What you see is what you get In the world of mechanism and chemical transformations Dr. Stephan Schürer ead of Intl. Sci. Content Libraria, Inc. sschurer@libraria.com Distribution of
5. Composition and Connectivity Does the formula always represent the complete composition of the substance?
 InChI FAQ InChI FAQ... 1 1. FAQ Overview... 5 1.1. What is this FAQ?... 5 1.2. Who is responsible for InChI?... 5 1.3. Where can I find out more?... 6 1.4. Is there an InChI mailing list?... 6 1.5. Are
InChI FAQ InChI FAQ... 1 1. FAQ Overview... 5 1.1. What is this FAQ?... 5 1.2. Who is responsible for InChI?... 5 1.3. Where can I find out more?... 6 1.4. Is there an InChI mailing list?... 6 1.5. Are
InChI, the IUPAC International Chemical Identifier
 eller et al. Journal of Cheminformatics (2015) 7:23 DI 10.1186/s13321-015-0068-4 RESEARC ARTICLE InChI, the IUPAC International Chemical Identifier Stephen R eller 1*, Alan Mcaught 2, Igor Pletnev 3, Stephen
eller et al. Journal of Cheminformatics (2015) 7:23 DI 10.1186/s13321-015-0068-4 RESEARC ARTICLE InChI, the IUPAC International Chemical Identifier Stephen R eller 1*, Alan Mcaught 2, Igor Pletnev 3, Stephen
Information Extraction from Chemical Images. Discovery Knowledge & Informatics April 24 th, Dr. Marc Zimmermann
 Information Extraction from Chemical Images Discovery Knowledge & Informatics April 24 th, 2006 Dr. Available Chemical Information Textbooks Reports Patents Databases Scientific journals and publications
Information Extraction from Chemical Images Discovery Knowledge & Informatics April 24 th, 2006 Dr. Available Chemical Information Textbooks Reports Patents Databases Scientific journals and publications
On InChI and evaluating the quality of cross-reference links
 Galgonek and Vondrášek Journal of Cheminformatics 2014, 6:15 RESEARCH ARTICLE Open Access On InChI and evaluating the quality of cross-reference links Jakub Galgonek * and Jiří Vondrášek * Abstract Background:
Galgonek and Vondrášek Journal of Cheminformatics 2014, 6:15 RESEARCH ARTICLE Open Access On InChI and evaluating the quality of cross-reference links Jakub Galgonek * and Jiří Vondrášek * Abstract Background:
AUTOMATIC GENERATION OF TAUTOMERS
 ПЛОВДИВСКИ УНИВЕРСИТЕТ ПАИСИЙ ХИЛЕНДАРСКИ БЪЛГАРИЯ НАУЧНИ ТРУДОВЕ, ТОМ 38, КН. 5, 2011 ХИМИЯ UNIVERSITY OF PLOVDIV PAISII HILENDARSKI BULGARIA SCIENTIFIC PAPERS, VOL. 38, BOOK 5, 2011 CHEMISTRY AUTOMATIC
ПЛОВДИВСКИ УНИВЕРСИТЕТ ПАИСИЙ ХИЛЕНДАРСКИ БЪЛГАРИЯ НАУЧНИ ТРУДОВЕ, ТОМ 38, КН. 5, 2011 ХИМИЯ UNIVERSITY OF PLOVDIV PAISII HILENDARSKI BULGARIA SCIENTIFIC PAPERS, VOL. 38, BOOK 5, 2011 CHEMISTRY AUTOMATIC
Introduction to Chemoinformatics
 Introduction to Chemoinformatics www.dq.fct.unl.pt/cadeiras/qc Prof. João Aires-de-Sousa Email: jas@fct.unl.pt Recommended reading Chemoinformatics - A Textbook, Johann Gasteiger and Thomas Engel, Wiley-VCH
Introduction to Chemoinformatics www.dq.fct.unl.pt/cadeiras/qc Prof. João Aires-de-Sousa Email: jas@fct.unl.pt Recommended reading Chemoinformatics - A Textbook, Johann Gasteiger and Thomas Engel, Wiley-VCH
Representation of molecular structures. Coutersy of Prof. João Aires-de-Sousa, University of Lisbon, Portugal
 Representation of molecular structures Coutersy of Prof. João Aires-de-Sousa, University of Lisbon, Portugal A hierarchy of structure representations Name (S)-Tryptophan 2D Structure 3D Structure Molecular
Representation of molecular structures Coutersy of Prof. João Aires-de-Sousa, University of Lisbon, Portugal A hierarchy of structure representations Name (S)-Tryptophan 2D Structure 3D Structure Molecular
Chemically Intelligent Experiment Data Management
 Chemically Intelligent Experiment Data Management Offering tools specifically designed to optimize the workflow of synthetic, medicinal, process and analytical chemists, the E-WorkBook Suite delivers a
Chemically Intelligent Experiment Data Management Offering tools specifically designed to optimize the workflow of synthetic, medicinal, process and analytical chemists, the E-WorkBook Suite delivers a
Marvin. Sketching, viewing and predicting properties with Marvin - features, tips and tricks. Gyorgy Pirok. Solutions for Cheminformatics
 Marvin Sketching, viewing and predicting properties with Marvin - features, tips and tricks Gyorgy Pirok Solutions for Cheminformatics The Marvin family The Marvin toolkit provides web-enabled components
Marvin Sketching, viewing and predicting properties with Marvin - features, tips and tricks Gyorgy Pirok Solutions for Cheminformatics The Marvin family The Marvin toolkit provides web-enabled components
Bioinformatics Workshop - NM-AIST
 Bioinformatics Workshop - NM-AIST Day 3 Introduction to Drug/Small Molecule Discovery Thomas Girke July 25, 2012 Bioinformatics Workshop - NM-AIST Slide 1/44 Introduction CMP Structure Formats Similarity
Bioinformatics Workshop - NM-AIST Day 3 Introduction to Drug/Small Molecule Discovery Thomas Girke July 25, 2012 Bioinformatics Workshop - NM-AIST Slide 1/44 Introduction CMP Structure Formats Similarity
So I have an SD File What do I do next? Rajarshi Guha & Noel O Boyle NCATS & NextMove So<ware
 So I have an SD File What do I do next? Rajarshi Guha & Noel O Boyle NCATS & NextMove Soonal Mee>ng, Boston 2015 What do you want to do? Tasks to be considered Searching for structures Managing
So I have an SD File What do I do next? Rajarshi Guha & Noel O Boyle NCATS & NextMove Soonal Mee>ng, Boston 2015 What do you want to do? Tasks to be considered Searching for structures Managing
CHAPTER 23 HW: ENOLS + ENOLATES
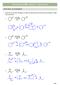 CAPTER 23 W: ENLS + ENLATES KET-ENL TAUTMERSM 1. Draw the curved arrow mechanism to show the interconversion of the keto and enol form in either trace acid or base. trace - 2 trace 3 + 2 + E1 2 c. trace
CAPTER 23 W: ENLS + ENLATES KET-ENL TAUTMERSM 1. Draw the curved arrow mechanism to show the interconversion of the keto and enol form in either trace acid or base. trace - 2 trace 3 + 2 + E1 2 c. trace
Reaxys Pipeline Pilot Components Installation and User Guide
 1 1 Reaxys Pipeline Pilot components for Pipeline Pilot 9.5 Reaxys Pipeline Pilot Components Installation and User Guide Version 1.0 2 Introduction The Reaxys and Reaxys Medicinal Chemistry Application
1 1 Reaxys Pipeline Pilot components for Pipeline Pilot 9.5 Reaxys Pipeline Pilot Components Installation and User Guide Version 1.0 2 Introduction The Reaxys and Reaxys Medicinal Chemistry Application
Dictionary of ligands
 Dictionary of ligands Some of the web and other resources Small molecules DrugBank: http://www.drugbank.ca/ ZINC: http://zinc.docking.org/index.shtml PRODRUG: http://www.compbio.dundee.ac.uk/web_servers/prodrg_down.html
Dictionary of ligands Some of the web and other resources Small molecules DrugBank: http://www.drugbank.ca/ ZINC: http://zinc.docking.org/index.shtml PRODRUG: http://www.compbio.dundee.ac.uk/web_servers/prodrg_down.html
Analyzing Small Molecule Data in R
 Analyzing Small Molecule Data in R Tyler Backman and Thomas Girke December 12, 2011 Analyzing Small Molecule Data in R Slide 1/49 Introduction CMP Structure Formats Similarity Searching Background Fragment
Analyzing Small Molecule Data in R Tyler Backman and Thomas Girke December 12, 2011 Analyzing Small Molecule Data in R Slide 1/49 Introduction CMP Structure Formats Similarity Searching Background Fragment
Chemical Journal Publishing in an Online World. Jason Wilde, Publisher Physical Sciences Nature Publishing Group ACS Spring Meeting 2009
 Chemical Journal Publishing in an Online World Jason Wilde, Publisher Physical Sciences Nature Publishing Group ACS Spring Meeting 2009 1 Overview Chemists and chemistry NPG and chemistry Nature Chemistry
Chemical Journal Publishing in an Online World Jason Wilde, Publisher Physical Sciences Nature Publishing Group ACS Spring Meeting 2009 1 Overview Chemists and chemistry NPG and chemistry Nature Chemistry
Comprehensive Chemoinformatics since Web-based, client/server, and toolkit approaches. Native Oracle (cartridge) and Microsoft technology.
 CambridgeSoft Solutions CambridgeSoft Research Informatics Louis Culot Executive Director, Research Informatics Division Informatics Overview ChemDraw since 1986. Comprehensive Chemoinformatics since 1998.
CambridgeSoft Solutions CambridgeSoft Research Informatics Louis Culot Executive Director, Research Informatics Division Informatics Overview ChemDraw since 1986. Comprehensive Chemoinformatics since 1998.
cheminformatics toolkits: a personal perspective
 cheminformatics toolkits: a personal perspective Roger Sayle Nextmove software ltd Cambridge uk 1 st RDKit User Group Meeting, London, UK, 4 th October 2012 overview Models of Chemistry Implicit and Explicit
cheminformatics toolkits: a personal perspective Roger Sayle Nextmove software ltd Cambridge uk 1 st RDKit User Group Meeting, London, UK, 4 th October 2012 overview Models of Chemistry Implicit and Explicit
Chemical Databases: Encoding, Storage and Search of Chemical Structures
 Chemical Databases: Encoding, Storage and Search of Chemical Structures Dr. Timur I. Madzhidov Kazan Federal University, Department of Organic Chemistry * Ray, L.C. and R.A. Kirsch, Finding Chemical Records
Chemical Databases: Encoding, Storage and Search of Chemical Structures Dr. Timur I. Madzhidov Kazan Federal University, Department of Organic Chemistry * Ray, L.C. and R.A. Kirsch, Finding Chemical Records
Reaction mechanisms offer us insights into how reactions work / how molecules react with one another.
 Introduction 1) Lewis Structures 2) Representing Organic Structures 3) Geometry and Hybridization 4) Electronegativities and Dipoles 5) Resonance Structures (a) Drawing Them (b) Rules for Resonance 6)
Introduction 1) Lewis Structures 2) Representing Organic Structures 3) Geometry and Hybridization 4) Electronegativities and Dipoles 5) Resonance Structures (a) Drawing Them (b) Rules for Resonance 6)
Supporting Information. Kekule.js: An Open Source JavaScript Chemoinformatics Toolkit
 Supporting Information Kekule.js: An Open Source JavaScript Chemoinformatics Toolkit Chen Jiang, *, Xi Jin, Ying Dong and Ming Chen Department of Organic Chemistry, China Pharmaceutical University, Nanjing
Supporting Information Kekule.js: An Open Source JavaScript Chemoinformatics Toolkit Chen Jiang, *, Xi Jin, Ying Dong and Ming Chen Department of Organic Chemistry, China Pharmaceutical University, Nanjing
Developing CAS Products for Substructure Searching by Chemists. Linda Toler
 Developing CAS Products for Substructure Searching by Chemists Linda Toler Developing CAS Products for Substructure Searching Evolution of the CAS Registry Development of substructure searching for CAS
Developing CAS Products for Substructure Searching by Chemists Linda Toler Developing CAS Products for Substructure Searching Evolution of the CAS Registry Development of substructure searching for CAS
California State Polytechnic University, Pomona. Exam Points 1. Nomenclature (1) 25
 hem 316 Final Spring, 2004 Beauchamp ame: Topic Total Points Exam Points 1. omenclature (1) 25 redit Tautomers (acidic or base conditions) 20 3. Reactions Page (10 x 3 = 30) 30 4. Aromatic Mechanism and
hem 316 Final Spring, 2004 Beauchamp ame: Topic Total Points Exam Points 1. omenclature (1) 25 redit Tautomers (acidic or base conditions) 20 3. Reactions Page (10 x 3 = 30) 30 4. Aromatic Mechanism and
A Journey from Data to Knowledge
 A Journey from Data to Knowledge Ian Bruno Cambridge Crystallographic Data Centre @ijbruno @ccdc_cambridge Experimental Data C 10 H 16 N +,Cl - Radspunk, CC-BY-SA CC-BY-SA Jeff Dahl, CC-BY-SA Experimentally
A Journey from Data to Knowledge Ian Bruno Cambridge Crystallographic Data Centre @ijbruno @ccdc_cambridge Experimental Data C 10 H 16 N +,Cl - Radspunk, CC-BY-SA CC-BY-SA Jeff Dahl, CC-BY-SA Experimentally
MEDICINAL CHEMISTRY I EXAM #1
 MEDICIAL CEMISTRY I EXAM #1 1 September 30, 2005 ame SECTI A. Answer each question in this section by writing the letter corresponding to the best answer on the line provided (2 points each; 50 points
MEDICIAL CEMISTRY I EXAM #1 1 September 30, 2005 ame SECTI A. Answer each question in this section by writing the letter corresponding to the best answer on the line provided (2 points each; 50 points
Хемоінформатика. Докінг. Дизайн ліків. Біоінформатика (3 курс) Лекція 4 (частина 1)
 Хемоінформатика. Докінг. Дизайн ліків Біоінформатика (3 курс) Лекція 4 (частина 1) Формати файлів в хемоінформатиці Chemical information is usually provided as files or streams and many formats have been
Хемоінформатика. Докінг. Дизайн ліків Біоінформатика (3 курс) Лекція 4 (частина 1) Формати файлів в хемоінформатиці Chemical information is usually provided as files or streams and many formats have been
CHE 321 Summer 2010 Exam 2 Form Choose the structure(s) that represent cis-1-sec-butyl-4-methylcyclohexane. I II III
 CE 321 Summer 2010 Exam 2 orm 2 Multiple Choice Questions: 60 points 1. Choose the structure(s) that represent cis-1-sec-butyl-4-methylcyclohexane. I II III (A) I only (B) II only (C) I and II only II
CE 321 Summer 2010 Exam 2 orm 2 Multiple Choice Questions: 60 points 1. Choose the structure(s) that represent cis-1-sec-butyl-4-methylcyclohexane. I II III (A) I only (B) II only (C) I and II only II
PubChem atom environments
 DOI 10.1186/s13321-015-0076-4 RESEARCH ARTICLE Open Access PubChem atom environments Volker D Hähnke, Evan E Bolton * and Stephen H Bryant Abstract Background: Atom environments and fragments find wide-spread
DOI 10.1186/s13321-015-0076-4 RESEARCH ARTICLE Open Access PubChem atom environments Volker D Hähnke, Evan E Bolton * and Stephen H Bryant Abstract Background: Atom environments and fragments find wide-spread
QSAR Modeling of Human Liver Microsomal Stability Alexey Zakharov
 QSAR Modeling of Human Liver Microsomal Stability Alexey Zakharov CADD Group Chemical Biology Laboratory Frederick National Laboratory for Cancer Research National Cancer Institute, National Institutes
QSAR Modeling of Human Liver Microsomal Stability Alexey Zakharov CADD Group Chemical Biology Laboratory Frederick National Laboratory for Cancer Research National Cancer Institute, National Institutes
Molecular Modelling. Computational Chemistry Demystified. RSC Publishing. Interprobe Chemical Services, Lenzie, Kirkintilloch, Glasgow, UK
 Molecular Modelling Computational Chemistry Demystified Peter Bladon Interprobe Chemical Services, Lenzie, Kirkintilloch, Glasgow, UK John E. Gorton Gorton Systems, Glasgow, UK Robert B. Hammond Institute
Molecular Modelling Computational Chemistry Demystified Peter Bladon Interprobe Chemical Services, Lenzie, Kirkintilloch, Glasgow, UK John E. Gorton Gorton Systems, Glasgow, UK Robert B. Hammond Institute
Synthetically Accessible Virtual Inventory (SAVI)
 Synthetically Accessible Virtual Inventory (SAVI) Yuri Pevzner, Wolf-Dietrich Ihlenfeldt 1, and Marc C. Nicklaus Computer-Aided Drug Design (CADD) Group 1 Xemistry GmbH, Germany Chemical Biology Laboratory
Synthetically Accessible Virtual Inventory (SAVI) Yuri Pevzner, Wolf-Dietrich Ihlenfeldt 1, and Marc C. Nicklaus Computer-Aided Drug Design (CADD) Group 1 Xemistry GmbH, Germany Chemical Biology Laboratory
Reaxys The Highlights
 Reaxys The Highlights What is Reaxys? A brand new workflow solution for research chemists and scientists from related disciplines An extensive repository of reaction and substance property data A resource
Reaxys The Highlights What is Reaxys? A brand new workflow solution for research chemists and scientists from related disciplines An extensive repository of reaction and substance property data A resource
Dock Ligands from a 2D Molecule Sketch
 Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Data Mining in the Chemical Industry. Overview of presentation
 Data Mining in the Chemical Industry Glenn J. Myatt, Ph.D. Partner, Myatt & Johnson, Inc. glenn.myatt@gmail.com verview of presentation verview of the chemical industry Example of the pharmaceutical industry
Data Mining in the Chemical Industry Glenn J. Myatt, Ph.D. Partner, Myatt & Johnson, Inc. glenn.myatt@gmail.com verview of presentation verview of the chemical industry Example of the pharmaceutical industry
Introduction Molecular Structure Script Console External resources Advanced topics. JMol tutorial. Giovanni Morelli.
 Gen 19th, 2017 1 2 Create and edit Display and view Mesurament and labelling Surface and Orbitals 3 4 from Database Protein Enzyme Crystal Structure and Unit Cell 5 Symmetry Animation General information
Gen 19th, 2017 1 2 Create and edit Display and view Mesurament and labelling Surface and Orbitals 3 4 from Database Protein Enzyme Crystal Structure and Unit Cell 5 Symmetry Animation General information
California State Polytechnic University, Pomona Chem 315. Exam Points 1. Nomenclature (1) 30
 hem 315 Final Exam Fall, 2007 Beauchamp ame: Topic Total Points Exam Points 1. omenclature (1) 30 redit 2. Relative rder of Reactivity 20 3. Reactions Page, using reactions learned thus far (30) 30 4.
hem 315 Final Exam Fall, 2007 Beauchamp ame: Topic Total Points Exam Points 1. omenclature (1) 30 redit 2. Relative rder of Reactivity 20 3. Reactions Page, using reactions learned thus far (30) 30 4.
ICM-Chemist How-To Guide. Version 3.6-1g Last Updated 12/01/2009
 ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
ICM-Chemist How-To Guide Version 3.6-1g Last Updated 12/01/2009 ICM-Chemist HOW TO IMPORT, SKETCH AND EDIT CHEMICALS How to access the ICM Molecular Editor. 1. Click here 2. Start sketching How to sketch
BIOVIA ENHANCED STEREOCHEMICAL REPRESENTATION WHITE PAPER
 BIOVIA ENHANCED STEREOCHEMICAL REPRESENTATION WHITE PAPER THE CHALLENGE Applied synthetic chemistry has been placing increasing emphasis in recent years on stereochemistry. In the pharmaceutical industry,
BIOVIA ENHANCED STEREOCHEMICAL REPRESENTATION WHITE PAPER THE CHALLENGE Applied synthetic chemistry has been placing increasing emphasis in recent years on stereochemistry. In the pharmaceutical industry,
Tautomerism in Alkyl and -OH Derivatives of Heterocycles containing Two Heteroatoms
 Tautomerism in Alkyl and - Derivatives of eterocycles containing Two eteroatoms Colette Jermann Master in chemistry - Year eterocyclic Chemistry Coursework 1 Summary Type of tautomerism S Isoxazol Isothiazole
Tautomerism in Alkyl and - Derivatives of eterocycles containing Two eteroatoms Colette Jermann Master in chemistry - Year eterocyclic Chemistry Coursework 1 Summary Type of tautomerism S Isoxazol Isothiazole
Chapter 19. Organic Chemistry. Carbonyl Compounds III. Reactions at the a-carbon. 4 th Edition Paula Yurkanis Bruice
 Organic Chemistry 4 th Edition Paula Yurkanis Bruice Chapter 19 Carbonyl Compounds III Reactions at the a-carbon Disampaikan oleh: Dr. Sri Handayani 2013 Irene Lee Case Western Reserve University Cleveland,
Organic Chemistry 4 th Edition Paula Yurkanis Bruice Chapter 19 Carbonyl Compounds III Reactions at the a-carbon Disampaikan oleh: Dr. Sri Handayani 2013 Irene Lee Case Western Reserve University Cleveland,
OAT Organic Chemistry - Problem Drill 19: NMR Spectroscopy and Mass Spectrometry
 OAT Organic Chemistry - Problem Drill 19: NMR Spectroscopy and Mass Spectrometry Question No. 1 of 10 Question 1. Which statement concerning NMR spectroscopy is incorrect? Question #01 (A) Only nuclei
OAT Organic Chemistry - Problem Drill 19: NMR Spectroscopy and Mass Spectrometry Question No. 1 of 10 Question 1. Which statement concerning NMR spectroscopy is incorrect? Question #01 (A) Only nuclei
O N N. electrons in ring
 ame I. ( points) Page 1 A. The compound shown below is a commonly prescribed antifungal drug. It belongs to a category of compounds known as triazoles. Analyze the geometry and other properties for various
ame I. ( points) Page 1 A. The compound shown below is a commonly prescribed antifungal drug. It belongs to a category of compounds known as triazoles. Analyze the geometry and other properties for various
CDK & Mass Spectrometry
 CDK & Mass Spectrometry October 3, 2011 1/18 Stephan Beisken October 3, 2011 EBI is an outstation of the European Molecular Biology Laboratory. Chemistry Development Kit (CDK) An Open Source Java TM Library
CDK & Mass Spectrometry October 3, 2011 1/18 Stephan Beisken October 3, 2011 EBI is an outstation of the European Molecular Biology Laboratory. Chemistry Development Kit (CDK) An Open Source Java TM Library
Exam 1 (Monday, July 6, 2015)
 Chem 231 Summer 2015 Assigned Homework Problems Last updated: Friday, July 24, 2015 Problems Assigned from Essential Organic Chemistry, 2 nd Edition, Paula Yurkanis Bruice, Prentice Hall, New York, NY,
Chem 231 Summer 2015 Assigned Homework Problems Last updated: Friday, July 24, 2015 Problems Assigned from Essential Organic Chemistry, 2 nd Edition, Paula Yurkanis Bruice, Prentice Hall, New York, NY,
Enolates, Enols, and Enamines Part 3
 Enolates, Enols, and Enamines Part 3 Guanine Tautomerize 2 2 Thymine C 3 Tautomerize C 3 Thursday March 21, 8:00-11:00 AM Final Exam Topics Part A: Carbonyl Fundamentals Carbonyl Survey Enolates, Enols,
Enolates, Enols, and Enamines Part 3 Guanine Tautomerize 2 2 Thymine C 3 Tautomerize C 3 Thursday March 21, 8:00-11:00 AM Final Exam Topics Part A: Carbonyl Fundamentals Carbonyl Survey Enolates, Enols,
Spring Term 2012 Dr. Williams (309 Zurn, ex 2386)
 Chemistry 242 Organic Chemistry II Spring Term 2012 Dr. Williams (309 Zurn, ex 2386) Web Page: http://math.mercyhurst.edu/~jwilliams/ jwilliams@mercyhurst.edu (or just visit Department web site and look
Chemistry 242 Organic Chemistry II Spring Term 2012 Dr. Williams (309 Zurn, ex 2386) Web Page: http://math.mercyhurst.edu/~jwilliams/ jwilliams@mercyhurst.edu (or just visit Department web site and look
The IUPAC InChI Chemical Structure Standard
 www.inchi-trust.org www.inchi-trust.org The IUPAC InChI Chemical Structure Standard Stephen Heller The main web sites for the IUPAC InChI project are: http://www.iupac.org/inchi and http://www.inchi-trust.org
www.inchi-trust.org www.inchi-trust.org The IUPAC InChI Chemical Structure Standard Stephen Heller The main web sites for the IUPAC InChI project are: http://www.iupac.org/inchi and http://www.inchi-trust.org
Chemistry /002 Exam 1 Version Green. The Periodic Table
 Name: Last First MI Chemistry 233-001/002 Exam 1 Version Green Fall 2018 Dr. J. sbourn Instructions: The first 13 questions of this exam should be answered on the provided Scantron. You must use a pencil
Name: Last First MI Chemistry 233-001/002 Exam 1 Version Green Fall 2018 Dr. J. sbourn Instructions: The first 13 questions of this exam should be answered on the provided Scantron. You must use a pencil
RESPONSE PROJECT DATABASE - NPS AND RELATED COMPOUNDS
 RESPONSE PROJECT DATABASE - NPS AND RELATED COMPOUNDS DESCRIPTION and GUIDELINES FOR USE Dr. Sonja Klemenc RS Ministry of the Interior, General Police Directorate National Forensic Laboratory Version 1.0
RESPONSE PROJECT DATABASE - NPS AND RELATED COMPOUNDS DESCRIPTION and GUIDELINES FOR USE Dr. Sonja Klemenc RS Ministry of the Interior, General Police Directorate National Forensic Laboratory Version 1.0
Isomerism. Introduction
 Isomerism Introduction The existence of two or more compounds with same molecular formula but different properties (physical, chemical or both) is known as isomerism; and the compounds themselves are called
Isomerism Introduction The existence of two or more compounds with same molecular formula but different properties (physical, chemical or both) is known as isomerism; and the compounds themselves are called
If somehow it is possible to increase the stability of a given base, then it
 Text Related to Segment.04 00 laude E. Wintner If somehow it is possible to increase the stability of a given base, then it should be true that the corresponding acid will be stronger than would be so
Text Related to Segment.04 00 laude E. Wintner If somehow it is possible to increase the stability of a given base, then it should be true that the corresponding acid will be stronger than would be so
Objective #1 (80 topics, due on 09/05 (11:59PM))
 Course Name: Chem 110 FA 2014 Course Code: N/A ALEKS Course: General Chemistry (First Semester) Instructor: Master Templates Course Dates: Begin: 06/27/2014 End: 06/27/2015 Course Content: 190 topics Textbook:
Course Name: Chem 110 FA 2014 Course Code: N/A ALEKS Course: General Chemistry (First Semester) Instructor: Master Templates Course Dates: Begin: 06/27/2014 End: 06/27/2015 Course Content: 190 topics Textbook:
Imago: open-source toolkit for 2D chemical structure image recognition
 Imago: open-source toolkit for 2D chemical structure image recognition Viktor Smolov *, Fedor Zentsev and Mikhail Rybalkin GGA Software Services LLC Abstract Different chemical databases contain molecule
Imago: open-source toolkit for 2D chemical structure image recognition Viktor Smolov *, Fedor Zentsev and Mikhail Rybalkin GGA Software Services LLC Abstract Different chemical databases contain molecule
Chap 11. Carbonyl Alpha-Substitution Reactions and Condensation Reactions
 Chap 11. Carbonyl Alpha-Substitution eactions and Condensation eactions Four fundamental reactions of carbonyl compounds 1) Nucleophilic addition (aldehydes and ketones) ) Nucleophilic acyl substitution
Chap 11. Carbonyl Alpha-Substitution eactions and Condensation eactions Four fundamental reactions of carbonyl compounds 1) Nucleophilic addition (aldehydes and ketones) ) Nucleophilic acyl substitution
Module No and Title. PAPER No: 5 ; TITLE : Organic Chemistry-II MODULE No: 25 ; TITLE: S E 1 reactions
 Subject Chemistry Paper No and Title Module No and Title Module Tag 5; Organic Chemistry-II 25; S E 1 reactions CHE_P5_M25 TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction 3. S E 1 reactions 3.1
Subject Chemistry Paper No and Title Module No and Title Module Tag 5; Organic Chemistry-II 25; S E 1 reactions CHE_P5_M25 TABLE OF CONTENTS 1. Learning Outcomes 2. Introduction 3. S E 1 reactions 3.1
Topic 9. Aldehydes & Ketones
 Chemistry 2213a Fall 2012 Western University Topic 9. Aldehydes & Ketones A. Structure and Nomenclature The carbonyl group is present in aldehydes and ketones and is the most important group in bio-organic
Chemistry 2213a Fall 2012 Western University Topic 9. Aldehydes & Ketones A. Structure and Nomenclature The carbonyl group is present in aldehydes and ketones and is the most important group in bio-organic
Navigation in Chemical Space Towards Biological Activity. Peter Ertl Novartis Institutes for BioMedical Research Basel, Switzerland
 Navigation in Chemical Space Towards Biological Activity Peter Ertl Novartis Institutes for BioMedical Research Basel, Switzerland Data Explosion in Chemistry CAS 65 million molecules CCDC 600 000 structures
Navigation in Chemical Space Towards Biological Activity Peter Ertl Novartis Institutes for BioMedical Research Basel, Switzerland Data Explosion in Chemistry CAS 65 million molecules CCDC 600 000 structures
The LSD Software. A tool for the structure determination of small molecules. Jean-Marc Nuzillard. Cargèse, 2013, March 23 th
 The LSD Software A tool for the structure determination of small molecules Jean-Marc Nuzillard University of Reims-Champagne-Ardenne Molecular Chemistry Institute Cargèse, 2013, March 23 th Small Molecules
The LSD Software A tool for the structure determination of small molecules Jean-Marc Nuzillard University of Reims-Champagne-Ardenne Molecular Chemistry Institute Cargèse, 2013, March 23 th Small Molecules
IUPAC International Chemical Identifier (InChI) Subcommittee
 IUPAC International Chemical Identifier (InChI) Subcommittee Minutes of the meeting on March 23rd 2009 at the Sheraton City Centre, Salt Lake City, UT, USA Present: Subcommittee members: Steve Heller (Chairman)
IUPAC International Chemical Identifier (InChI) Subcommittee Minutes of the meeting on March 23rd 2009 at the Sheraton City Centre, Salt Lake City, UT, USA Present: Subcommittee members: Steve Heller (Chairman)
Navigating between patents, papers, abstracts and databases using public sources and tools
 Navigating between patents, papers, abstracts and databases using public sources and tools Christopher Southan 1 and Sean Ekins 2 TW2Informatics, Göteborg, Sweden, Collaborative Drug Discovery, North Carolina,
Navigating between patents, papers, abstracts and databases using public sources and tools Christopher Southan 1 and Sean Ekins 2 TW2Informatics, Göteborg, Sweden, Collaborative Drug Discovery, North Carolina,
C. Correct! The abbreviation Ar stands for an aromatic ring, sometimes called an aryl ring.
 Organic Chemistry - Problem Drill 05: Drawing Organic Structures No. 1 of 10 1. What does the abbreviation Ar stand for? (A) Acetyl group (B) Benzyl group (C) Aromatic or Aryl group (D) Benzoyl group (E)
Organic Chemistry - Problem Drill 05: Drawing Organic Structures No. 1 of 10 1. What does the abbreviation Ar stand for? (A) Acetyl group (B) Benzyl group (C) Aromatic or Aryl group (D) Benzoyl group (E)
The Fragment Network: A Chemistry Recommendation Engine Built Using a Graph Database
 Supporting Information The Fragment Network: A Chemistry Recommendation Engine Built Using a Graph Database Richard J. Hall, Christopher W. Murray and Marcel L. Verdonk. Contents This supporting information
Supporting Information The Fragment Network: A Chemistry Recommendation Engine Built Using a Graph Database Richard J. Hall, Christopher W. Murray and Marcel L. Verdonk. Contents This supporting information
Chemistry 233 Exam 3. The Periodic Table
 Name: Last First MI Chemistry 233 Exam 3 Fall 2017 Dr. J. sbourn Instructions: The first 8 questions of this exam should be answered on the provided Scantron. You must use a pencil for filling in the Scantron
Name: Last First MI Chemistry 233 Exam 3 Fall 2017 Dr. J. sbourn Instructions: The first 8 questions of this exam should be answered on the provided Scantron. You must use a pencil for filling in the Scantron
The following exam contains 30 questions valued at 3 point/question and bonus opportunities. Name:
 Chemistry 261 Exam 3 all 2017 The following exam contains 30 questions valued at 3 point/question and bonus opportunities Name: ELECTROPILIC ADDITION REACTIONS BASIC REACTION OUTCOMES 1. Addition of hydrogen
Chemistry 261 Exam 3 all 2017 The following exam contains 30 questions valued at 3 point/question and bonus opportunities Name: ELECTROPILIC ADDITION REACTIONS BASIC REACTION OUTCOMES 1. Addition of hydrogen
Structure Searching in CrossFire Beilstein. DiscoveryGate SM Version 1.4 Participant s Guide
 Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Elsevier MDL 14600
Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Structure Searching in CrossFire Beilstein DiscoveryGate SM Version 1.4 Participant s Guide Elsevier MDL 14600
CHEM 341: Organic Chemistry I at North Dakota State University Midterm Exam 01 - Fri, Feb 10, 2012!! Name:!
 CEM 341: rganic Chemistry I at North Dakota State University Midterm Exam 01 - Fri, Feb 10, 2012!! Name:! Please read through each question carefully and answer in the spaces provided. A good strategy
CEM 341: rganic Chemistry I at North Dakota State University Midterm Exam 01 - Fri, Feb 10, 2012!! Name:! Please read through each question carefully and answer in the spaces provided. A good strategy
Style guide for chemical structures
 Style guide for chemical structures Although there are a number of ways that chemical structures can be drawn based on individual preferences, our journals aim to use consistent styling wherever possible.
Style guide for chemical structures Although there are a number of ways that chemical structures can be drawn based on individual preferences, our journals aim to use consistent styling wherever possible.
Computational Chemistry in Drug Design. Xavier Fradera Barcelona, 17/4/2007
 Computational Chemistry in Drug Design Xavier Fradera Barcelona, 17/4/2007 verview Introduction and background Drug Design Cycle Computational methods Chemoinformatics Ligand Based Methods Structure Based
Computational Chemistry in Drug Design Xavier Fradera Barcelona, 17/4/2007 verview Introduction and background Drug Design Cycle Computational methods Chemoinformatics Ligand Based Methods Structure Based
UniChem: extension of InChI-based compound mapping to salt, connectivity and stereochemistry layers
 Chambers et al. Journal of Cheminformatics 2014, 6:43 DATABASE Open Access UniChem: extension of InChI-based compound mapping to salt, connectivity and stereochemistry layers Jon Chambers *, Mark Davies,
Chambers et al. Journal of Cheminformatics 2014, 6:43 DATABASE Open Access UniChem: extension of InChI-based compound mapping to salt, connectivity and stereochemistry layers Jon Chambers *, Mark Davies,
Open PHACTS Explorer: Compound by Name
 Open PHACTS Explorer: Compound by Name This document is a tutorial for obtaining compound information in Open PHACTS Explorer (explorer.openphacts.org). Features: One-click access to integrated compound
Open PHACTS Explorer: Compound by Name This document is a tutorial for obtaining compound information in Open PHACTS Explorer (explorer.openphacts.org). Features: One-click access to integrated compound
BioSolveIT. A Combinatorial Approach for Handling of Protonation and Tautomer Ambiguities in Docking Experiments
 BioSolveIT Biology Problems Solved using Information Technology A Combinatorial Approach for andling of Protonation and Tautomer Ambiguities in Docking Experiments Ingo Dramburg BioSolve IT Gmb An der
BioSolveIT Biology Problems Solved using Information Technology A Combinatorial Approach for andling of Protonation and Tautomer Ambiguities in Docking Experiments Ingo Dramburg BioSolve IT Gmb An der
Lecture 13A 05/11/12. Amines. [Sn2; Hofmann elimination; reduction of alkyl azides, amides, nitriles, imines; reductive amination; Gabriel synthesis]
![Lecture 13A 05/11/12. Amines. [Sn2; Hofmann elimination; reduction of alkyl azides, amides, nitriles, imines; reductive amination; Gabriel synthesis] Lecture 13A 05/11/12. Amines. [Sn2; Hofmann elimination; reduction of alkyl azides, amides, nitriles, imines; reductive amination; Gabriel synthesis]](/thumbs/86/93838793.jpg) Lecture 13A 05/11/12 Amines [Sn2; ofmann elimination; reduction of alkyl azides, amides, nitriles, imines; reductive amination; Gabriel synthesis] Curtius and ofmann rearrangements Both of these, in principle,
Lecture 13A 05/11/12 Amines [Sn2; ofmann elimination; reduction of alkyl azides, amides, nitriles, imines; reductive amination; Gabriel synthesis] Curtius and ofmann rearrangements Both of these, in principle,
Web-accessible Chemical. Compound Information. Dana L. Roth
 Web-accessible Chemical Compound Information Dana L. Roth ABSTRACT. Web-accessible chemical compound information resources are widely available. In addition to fee-based resources, such as SciFinder Scholar
Web-accessible Chemical Compound Information Dana L. Roth ABSTRACT. Web-accessible chemical compound information resources are widely available. In addition to fee-based resources, such as SciFinder Scholar
The Electronic Representation of Chemical Structures: beyond the low hanging fruit
 The Electronic Representation of Chemical Structures: beyond the low hanging fruit How Accelrys Plans to Address the Remaining Challenges in Structure Representation and Searching: Chemically Modified
The Electronic Representation of Chemical Structures: beyond the low hanging fruit How Accelrys Plans to Address the Remaining Challenges in Structure Representation and Searching: Chemically Modified
CHEM 240: Survey of Organic Chemistry at North Dakota State University Midterm Exam 02 - Tue, 23 Sep 2014!! Name:! KEY!
 CEM 240: Survey of rganic Chemistry at orth Dakota State University Midterm Exam 02 - Tue, 23 Sep 2014!! ame:! KEY! Please read through each question carefully and answer in the spaces provided. A good
CEM 240: Survey of rganic Chemistry at orth Dakota State University Midterm Exam 02 - Tue, 23 Sep 2014!! ame:! KEY! Please read through each question carefully and answer in the spaces provided. A good
C h a p t e r T w e n t y - o n e : Enols, Enolates, and Aldol-like Condensations
 h a p t e r T w e n t y o n e : Enols, Enolates, and Aldollike ondensations Li LDA 1. R TF R 2. 3 R A directed aldol reaction, used in the synthesis of periplanone B, a cockroach attractant M 323: Summary
h a p t e r T w e n t y o n e : Enols, Enolates, and Aldollike ondensations Li LDA 1. R TF R 2. 3 R A directed aldol reaction, used in the synthesis of periplanone B, a cockroach attractant M 323: Summary
CHEM1101 Worksheet 6: Lewis Structures
 CHEM1101 Worksheet 6: Lewis Structures Model 1: Simple Compounds of C, N, O and F The octet rule tells us that C, N, O and F will form covalent bonds so that they are surrounded by eight electrons. For
CHEM1101 Worksheet 6: Lewis Structures Model 1: Simple Compounds of C, N, O and F The octet rule tells us that C, N, O and F will form covalent bonds so that they are surrounded by eight electrons. For
CEM 850 Final Exam H N H H O H H O H C H 2. MeO
 CEM 850 Final Exam December 11, 2003 This exam consists of 20 pages. Be sure to write your name on all pages of the exam. Please write legibly and draw all structures clearly. Good luck. 1. (10 pts) Estimate
CEM 850 Final Exam December 11, 2003 This exam consists of 20 pages. Be sure to write your name on all pages of the exam. Please write legibly and draw all structures clearly. Good luck. 1. (10 pts) Estimate
Fri 6 Nov 09. More IR Mass spectroscopy. Hour exam 3 Fri Covers Chaps 9-12 Wednesday: Review
 Fri 6 Nov 09 our exam 3 Fri 11-13 Covers Chaps 9-12 Wednesday: Review More IR Mass spectroscopy Good web site for IR, Mass, NMR spectra: http://riodb01.ibase.aist.go.jp/sdbs/cgi-bin/cre_index.cgi?lang=eng
Fri 6 Nov 09 our exam 3 Fri 11-13 Covers Chaps 9-12 Wednesday: Review More IR Mass spectroscopy Good web site for IR, Mass, NMR spectra: http://riodb01.ibase.aist.go.jp/sdbs/cgi-bin/cre_index.cgi?lang=eng
Final Exam. Chem 3B, Fall 2016 Monday, Dec 12, pm. Name Answer Key. Student ID. If you are making up an incomplete, list the semester here:
 Final Exam Chem 3B, Fall 2016 Monday, Dec 12, 2016 3-6 pm ame Answer Key Student ID If you are making up an incomplete, list the semester here: You have 180 minutes to complete this exam. Please provide
Final Exam Chem 3B, Fall 2016 Monday, Dec 12, 2016 3-6 pm ame Answer Key Student ID If you are making up an incomplete, list the semester here: You have 180 minutes to complete this exam. Please provide
For more info visit
 Bond Fission: a) Homolytic fission: Each atom separates with one electron, leading to the formation of highly reactive entities called radicals, owing their reactivity to their unpaired electron. b) Heterolytic
Bond Fission: a) Homolytic fission: Each atom separates with one electron, leading to the formation of highly reactive entities called radicals, owing their reactivity to their unpaired electron. b) Heterolytic
Keynotes in Organic Chemistry
 Keynotes in Organic Chemistry Second Edition ANDREW F. PARSONS Department of Chemistry, University of York, UK Wiley Contents Preface xi 1 Structure and bonding 1 1.1 Ionic versus covalent bonds 1 1.2
Keynotes in Organic Chemistry Second Edition ANDREW F. PARSONS Department of Chemistry, University of York, UK Wiley Contents Preface xi 1 Structure and bonding 1 1.1 Ionic versus covalent bonds 1 1.2
Chemistry 233 Exam 3 (Green) The Periodic Table
 Name: Last First MI Chemistry Exam (Green) Summer 7 Dr. J. sbourn Instructions: The first questions of this exam should be answered on the provided Scantron. You must use a pencil for filling in the Scantron
Name: Last First MI Chemistry Exam (Green) Summer 7 Dr. J. sbourn Instructions: The first questions of this exam should be answered on the provided Scantron. You must use a pencil for filling in the Scantron
Metrabase The Metabolism and Transport Database. user manual v
 Metrabase The Metabolism and Transport Database user manual v2015.02 Contents 1 INTRODUCTION 3 2 METRABASE CONTENT 4 2.1 ACTIVITIES (INTERACTIONS) 7 2.2 PROTEINS 9 2.3 COMPOUNDS 10 2.4 DATA SOURCES 13
Metrabase The Metabolism and Transport Database user manual v2015.02 Contents 1 INTRODUCTION 3 2 METRABASE CONTENT 4 2.1 ACTIVITIES (INTERACTIONS) 7 2.2 PROTEINS 9 2.3 COMPOUNDS 10 2.4 DATA SOURCES 13
Organic Chemistry. Alkynes
 For updated version, please click on http://ocw.ump.edu.my Organic Chemistry Alkynes by Dr. Seema Zareen & Dr. Izan Izwan Misnon Faculty Industrial Science & Technology seema@ump.edu.my & iezwan@ump.edu.my
For updated version, please click on http://ocw.ump.edu.my Organic Chemistry Alkynes by Dr. Seema Zareen & Dr. Izan Izwan Misnon Faculty Industrial Science & Technology seema@ump.edu.my & iezwan@ump.edu.my
Virtual Libraries and Virtual Screening in Drug Discovery Processes using KNIME
 Virtual Libraries and Virtual Screening in Drug Discovery Processes using KNIME Iván Solt Solutions for Cheminformatics Drug Discovery Strategies for known targets High-Throughput Screening (HTS) Cells
Virtual Libraries and Virtual Screening in Drug Discovery Processes using KNIME Iván Solt Solutions for Cheminformatics Drug Discovery Strategies for known targets High-Throughput Screening (HTS) Cells
Basic Techniques in Structure and Substructure
 Truncating Molecules Basic Techniques in Structure and Substructure Searching for Information Professionals Judith Currano Head, Chemistry Library University of Pennsylvania currano@pobox.upenn.edu Acknowledgements
Truncating Molecules Basic Techniques in Structure and Substructure Searching for Information Professionals Judith Currano Head, Chemistry Library University of Pennsylvania currano@pobox.upenn.edu Acknowledgements
The IUPAC Chemical Identifier Technical Manual
 The IUPAC Chemical Identifier Technical Manual Stephen E. Stein, Stephen R. eller, Dmitrii V. Tchekhovskoi Physical and Chemical Properties Division ational Institute of Standards and Technology Gaithersburg,
The IUPAC Chemical Identifier Technical Manual Stephen E. Stein, Stephen R. eller, Dmitrii V. Tchekhovskoi Physical and Chemical Properties Division ational Institute of Standards and Technology Gaithersburg,
BioSolveIT. A Combinatorial Docking Approach for Dealing with Protonation and Tautomer Ambiguities
 BioSolveIT Biology Problems Solved using Information Technology A Combinatorial Docking Approach for Dealing with Protonation and Tautomer Ambiguities Ingo Dramburg BioSolve IT Gmb An der Ziegelei 75 53757
BioSolveIT Biology Problems Solved using Information Technology A Combinatorial Docking Approach for Dealing with Protonation and Tautomer Ambiguities Ingo Dramburg BioSolve IT Gmb An der Ziegelei 75 53757
Tautomer Identification and Tautomer Structure Generation Based on the InChI Code
 J. Chem. Inf. Model. 2010, 50, 1223 1232 1223 Tautomer Identification and Tautomer Structure Generation Based on the InChI Code Torsten Thalheim,, Armin Vollmer, Ralf-Uwe Ebert, Ralph Kühne, and Gerrit
J. Chem. Inf. Model. 2010, 50, 1223 1232 1223 Tautomer Identification and Tautomer Structure Generation Based on the InChI Code Torsten Thalheim,, Armin Vollmer, Ralf-Uwe Ebert, Ralph Kühne, and Gerrit
