SDS-PAGE Electrophoresis
|
|
|
- Alexander Mark Davidson
- 6 years ago
- Views:
Transcription
1 SDS-PAGE Electrophoresis Anirbit Department of Theoretical Physics, Tata Institute of Fundamental Research, Mumbai , India (Dated: February 15, 2010) In this report I shall explain the steps involved in doing an electrophoresis experiment on proteins and eventual analysis of the data to study how best one can predict the mass of a protein from the intensity of the dye stains. The particular version of electrophoresis that will be used is called SDS-PAGE in literature because of the essential use of the chemical Sodium Dodecyle Sulphate and Polyacrylamide in the process. En route one shall unravel many subtleties of the prediction process and realize the intrinsic problems of doing a simple intensity-mass plots as compared to more robust power law behaviour that will be be seen to emerge in variation of intensity with logarithm of fractional masses. I. INTRODUCTION To start off one can imagine the electrophoresis as a method by which a solution of proteins is passed through a gel in the presence of a constant potential difference such that the proteins of various sizes flow through it at different speeds and hence one can tell the constituents of the solution based on the positions of a dye stain which binds to the proteins. In the future experiments that will be done in this laboratory it will be important that one can monitor fractional changes of a certain protein during the run of an experiment. In most of these proteins knowing the exact amount of protein in the solution is very hard. In this experiment the attempt shall be to try to extend the applicability of electrophoresis to see if given a pure protein solution one can predict the total mass of protein in it by correlating the intensity of the dye stain with the mass of the protein. II. SDS-PAGE ELECTROPHORESIS In the given electrophoresis set-up of Bio-Rad one can set up 2 gels to run simultaneously. One prepares the solutions of chemicals in volumes slightly larger than what is required to account for the spill-overs that are very common. To run 2 gels together I found it convenient to make 15ml of 8% solution for Separating Gel and 5ml of solution for Stacking Gel. FIG. 1: The upper part (the darker part after staining) of the gel is called the Stacking Gel and the lower part is called the Separating Gel. There is also a conducting electrolyte in the system which completes the circuit externally between these two parts of the gel.
2 2 A. A note on the chemical solutions used Some of the standard acronyms for chemicals that are used are, Acrylamide Mix is a 1 : 29 mixture of N, Nmethylene bis-acrylamide and Acrylamide SDS is Sodium Dodecyl Sulphate APS is Ammonium per-sulphate (always stored at 20 C) TEMED is N,N,N,N -Tetramethylethyl-enediamine (always stored at 4 C) All the autoclaving in this experiment are done at 121 C for 15minutes at 103kP a of pressure in an autoclaving apparatus made by Hospharma. The mixture of chemicals required to make 15ml of Separating Gel are, H 2 O 6.9ml 30%Acrylamide M ix 4.0ml 1.5M T ris Hcl(pH 8.8) 3.8ml 10% SDS 0.15ml 10% AP S 0.15ml T EMED 0.009ml The mixture of chemicals required to make 5ml of Stacking Gel are, H 2 O 3.4ml 30%Acrylamide M ix 0.83ml 1M T ris Hcl(pH 8.8) 0.63ml 10% SDS 0.05ml 10% AP S 0.05ml T EMED 0.005ml To make 2l of the Electrophoresis Buffer that will act as the conductor for the current during electrophoresis one uses the following mixture, Trizma Base (a.k.a Tris), which is mainly C 4 H 11 NO 3 12g Glycine 57.6g SDS 2g Autoclaved water is added to make up the volume to 2l and ph adjustment is not done. To make 50ml of the Sample Buffer (DTT dye) one has to mix the following, 0.6M solution of Tris in HCl at ph 6.8 5ml SDS 0.5g Sucrose 5g β Mercaptoethanol 0.25ml 0.5% Bromophenol Blue 5ml Autoclaved water is added to make up the solution to 50ml
3 3 To make Staining Solution one mixes, 0.1%(v/v) Coomassie brilliant blue R250 50%(v/v) Methanol 10%(v/v) Glacial Acetic acid. To make the Destaining Solution one mixes, 10% Methanol 7% Glacial Acetic acid To make many of these solutions the masses of the chemicals needed to be measured and they were done on these two machines manufactured by Sartorius, One was the model CP 1245 and it had a maximum load capacity of 120g with a least count of 10 4 g The other was of model BL1500S and it had a maximum load capacity of 1500g with a least count of 10 2 g The following are the steps in setting the gels, B. Setting the gels Before one starts making the solutions one sets up the two pairs of glass plates on the rack in a particular orientation as marked on the plates. Each of these glass plates is about 1mm think and specially made to be as smooth as possible and are very fragile. The larger plat is of dimensions about 10cm 8.2cm and the smaller one is of size about 10cm 7.3cm. There are two about 1mm thick strips of opaque glass of sizes almost 8.2cm 9mm stuck on the two sides of the larger glass. This ensures a gap of 1mm between the two glass plates in which the gel will be polymerized. One makes the solutions of the separating and the stacking gel in separate containers. I made them in discarded 50ml aliquots manufactured by Falcon. Except APS and TEMED one mixes in all the chemicals in the two compositions given above. APS needs to be freshly prepared or one that has been stored at 20 C and just taken out. Since the separating gel will be made first hence for now one puts the APS in the separating solution only and mixes the TEMED also in only that one. As soon as the APS and TEMED are poured into the separating solution one immediately transfers them between one of the pairs of glass-plates through a pipette tip and the tip should be moved all along the edge through out to ensure that the fluid flows in uniformly between the glass plates. Any non-uniformity will result in nucleation centers for the polymerization process and make the gel non-homogeneous. One stops pouring the solution when it reaches an height of about 1cm below the edge of the plates (such that the gap is larger than the depth of the comb so that the protein solutions have to pass through some stacking gel before entering the separating gel andin the process they undergo isoelectric focussing through the process of isotacphoresis.) One pours butanol on the top of it through another pipette. This is immiscible with the rest of the solution and floats on the top helping to smoothen the gel surface and also acts as a protecting layer from the oxidation effects of the atmosphere. Now one waits for about 30mins for the gel to form. It can sometimes take an hour or a little more depending on humidity and temperature conditions. After about 30mins I keep checking for the formation of the gel by tilting the whole set up at times to see if the butanol layer is hitting a hard boundary on its lower surface. Once the separating gel is formed one washes out the butanol layer by tilting the container and gently flowing in autoclaved water through the gap and absorbing any further remnant butanol by gently inserting a tissue paper in the gap.
4 4 FIG. 2: The above photograph shows green frames in which the gel is set and the transparent rack in which the green frames are clamped to keep them stable during the polymerization process. Now one mixes the APS and the TEMED in the stacking solution and pours it into the gap between the pairs of plates through a pipette in a similar fashion. Now one has to quickly insert the comb into the gap which will demarcate the wells that will form inside the stacking gel. This comb is not to be taken out untill the protein loading starts. This set up should be left undisturbed till it is necessary to install the gels into the electrophoresis tank. Generally during this time one starts making the chemicals that will be loaded into the gel. Like the dilution series of Actin and BSA to be described below. C. Making Actin dilution solution When Actin is extracted it is not known as to how much Actin is present in the solution. It is estimated from indirect methods that the concentration is 2µg/mul. The various steps in preparing the Actin dilution are as follows, I took 5 aliquots of the Actin solution which gave me about 43µl of Actin solution. I added 117µl of H2 O to it and hence made up the solution to 160µl. Call this solution A0 I took 80µl of A0 and added 80µl of H2 O to it and made 160µl of another solution. Lets name it A1 Again I took 80µl of A1 and added 80µl of H2 O to it and made 160 of another solution. Lets name it A2. Thus iterating the process about 5 times one makes solutions of 80µl each till A5. In the making of A5 the 80µl of A4 that remains is preserved separately for future experiments and not used in the dyeing and heating in the next steps. In each of the 80µl solutions from A0 to A5 of Actin, 20µl of the 5X DTT dye is added (so that in the final solution the dye is in concentration of 1X ) and then each of these 100µl solutions is sent for heating for 5mins at 100 C. Post heating one loads 20µl of each solution in each of the wells of the gel. Hence the amounts of protein in the wells of the gel fall in inverse powers of two.
5 5 D. Making BSA dilution solution The steps to prepare the BSA sample that will be loaded in the gel, 1. 5mg of BSA is mixed in 1ml of water. (call this solution A 0 ) ml of A 0 is taken in another aliquot and 0.5ml of water is mixed in it (call it solution A 1 ) ml of A 1 is taken in another aliquot and 0.5mk of water is mixed in it (call it solution A 2 ). 4. In this way solutions till A 5 are made µl of A 0 is taken in an aliquot and 7.5µl of the 5X dye is mixed in it. (This choice is explained in the last point) 6. So the aliquout now contains 150µg of BSA in a solution of volume 37.5µl. 7. To denature the proteins boil this 37.5µl of solution for 5min at 100 C (call this B 0 ) 8. The steps 5,6 and 7 are repeated for A 1, A 2, A 3, A 4, A 5 and call the corresponding 37.5µl solutions B 1, B 2, B 3, B 4, B µl of each B i is taken and loaded into each of the wells. 10. Then the mass of the BSA loaded in each of the wells are respectively 12µg, 6µg, 3µg, 1.5µg, 0.75µg and 0.375µg. 11. The 5X dye is a solution of the dye made and stored in such a way that it has concentration 5 times more than what it should be in the final solution. Then one can see that adding 7.5µl of such a 5X dye to 30µl of fluids results in the correct concentration of the dye in the final mixture. The steps are as follows, E. Loading, running, staining and destaining the gel One generally needs to have about 500ml of the electrophoresis buffer as a conductor for running the gel. 50ml of the available 10X buffer is mixed with 450ml of autoclaved water to make the required solution. First the two glass framed gels are slid into the slots on the two sides of the frame that goes into the tank. The glass frames are positioned so that the shorter plate faces inward. Checking for snug fitting one locks them by the plastic hinges on the frame. Then the frame is inserted into the tank and the electrophoresis buffer is poured into the volume between the two glass frames. One checks if the buffer is leaking out into the tank. If it does then one has to diamantle the frame and reset it again. Then one pours the rest of the prepared buffer in the volume surrounding the frame inside the tank. Now one should pull out the combs taking care that the jerk does not break the wells inside the gel that would have formed. At certain angles the wells will be visibile to the naked eye because of the refractive index difference between air and the transparent gel. To reduce chances of loading the protein at regions other than the gel one can mark the positions of the wells on the wall of the tank using some marker. The proteins are loaded into the micropipettes from the aliquots and the pippete is lightly inserted into the well from the side of the shorter glass plate and the protein pushed in. One can see the coloured protein settled into the well. Care should be taken to push in the protein in one shot since doing a suction can contaminate the protein by the buffer solution. After all the gels are loaded, the top cover is closed and it is connected to the D.C source that comes with the instrument. It is generally run at about 100V. This D.C source has ratings of delivering 2.5A current at 500V delivering a maximum power of 500W.
6 6 FIG. 3: The above photograph shows the electrophoresis set-up while it is running. FIG. 4: The top-view of the electrophoresis set-up while it is running.
7 7 The voltage is maintained till the blue line in the gel (formed due to the accumulation of the dye) hits the lower edge of the gel. As a thumb rule the protein is generally behind dye. One shouldn t leave the process unattended for long at this stage since the proteins can very well run out of the gel into the solution if kept for long. FIG. 5: The faint blue line towards the bottom of the gel indicating the distance through which the dye has run. After the gel running is complete, the apparantus is dismantled and the glass plates taken out. The two glass plates in every pair will stick to each other very strongly because of surface tension and just trying to pull them apart runs the risk of breaking the fragile glass and also the gel. In general water is lighly run through the upper edge and then softly one can pry them apart using a plastic wedge. Using the same wedge the gel is lighly detached from the glass plate and this thin film that is formed is transferred to a tray. Now one can either use this gel to do staining with Coomassie blue dye or use it for Western Blotting. The next steps of Coomassie blue staining are relatively simple which are listed below and Western Blotting is a longer sequence of steps which was unsuccessfully tried and shall not be explained here. The steps of staining and destaining are as follows, On the gels which are transferred into the tray, Coomassie Blue dye solution is poured and the set up is put on a rocker for overnight staining. This is staining is generally done for about 10 12hours and during this time the oscillations of the rocker ensure that the dye keeps flowing across the gel uniformly without clustering anywhere and hence stains the gel uniformly. After the staining is over the remaining staining solution is recovered for reuse. The destaining solution is is then pourd in and set on a rocker. As the stain dissolves into the destaining solution the solution will keep becoming blue and then one decants out the solution and pours in fresh solution. This change has to be done almost every minutes for about 5hours. During this destaining process care must be taken not to overdo the process since it can eventually start dissolving out the dye stuck to the protein bands also. Initially the gel looks uniformly blue after about 2 rounds of destaining the protein bands become visible in contrast with the lighter background.
8 8 Some general dimensions of the gel are, F. Some measurements of the gel The separating gel is generally about 5.5cm 8.2cm in size and the stacking gel is 1.5cm 8.2cm with wells along one edge. The wells in the stacking gel are about 1cm deep and 0.5cm in width, so the proteins pass through about 0.6cm of stacking gel before hitting the separating gel. The gel is generally 0.1cm in thickness. The separating gel is about 4.26g in mass and is typically at a density of 0.91g/cc. The stacking gel is about 0.83g in mass and is typically at a density of 1.03g/cc G. A short note about the chemicals The precise utility of each of the chemicals in the experiment is a extensive question in chemistry and the rational behind the quatities of each to be used is hard to theoretically justiy. Some of the essential physical ideas that go behind it shall be explained at the end in the section on background theory. A basic list of use of the important chemicals are, Sodium Dodecyle Sulphate is basically a soap which when heated with the protein binds to the amino acids in the protein and hence destabilizes the hydrogen-bonding which renders the protein its structure. This denatures the protein by making all of them more or less cylindrical in shape. This ensures that the proteins s mobility which determines their stacking sequence depends less on the structure and more on the molecular mass. Also, anions of SDS bind to the main peptide chain at a ratio of one SDS anion for every two amino acid residues. This effectively imparts a negative charge on the protein that is proportional to the mass of that protein (about 1.4gSDS/gprotein). This negative charging is essential for the migration of the proteins (precisely the protein- SDS complex) towards the anode during electrophoresis. FIG. 6: SDS β Mercaptoethanol, HO (CH 2 ) 2 SH is mixed in the protein solution mainly to break the di-sulphide bonds in the protein which also help keep the protein conformation.
9 9 FIG. 7: The basic idea is of two amino acids bonding to form a peptide. This will exist mostly as a zwitterion in solution and SDS when heated at 100 C for 5 minutes with proteins will bind at the rate of one SDS anion for every two amino acid residues or approximately as 1.4gSDS/g protein. This not only breaks the hydrogen bonding responsible for the protein conformation but will also give a net negative charge to the protein essential for its migration towards the anode during electrophoresis. FIG. 8: The above is the typical reaction by which β Mercaptoethanol cleaves the disulphide bond in proteins The steps in the polymerization are as follows, TEMED catalyzes the decomposition of the persulphate ion S to give a free radical as, S e 1 SO4 2 + SO4 1 If M is the acylamide monomer then polymerization proceeds by iterating the following, R + M RM RM + M RMM RMM + M RMM The dissolved 0 2 mops up the free-radical. and so on...
10 The Glycine provides the ion with the slowest mobility which helps in stacking the proteins during the transport in the presence of the electric field. The Chloride ions provide the background conducting medium and is the leading ion being the ion around with the highest mobility. Hence the proteins being of intermediate mobility get stacked between the chloride and the glycine ions. Bromophenol Blue is employed tracking dye mixed in the protein solution, because it is viable in alkali and neutral ph, it is a small molecule, it is negatively charged above ph 4.6 and hence moves towards the anode. Being a small molecule it moves ahead of most proteins and nucleic acids. As it reaches the anodic end of the electrophoresis medium electrophoresis is stopped. It can bind with proteins weakly and give blue colour. 10 FIG. 9: Structure of Bromophenol Blue Coomassie Blue R-250 is an anionic dye used in the staining solution.this binds with proteins non-specifically. As SDS is also anionic, it may interfere with staining process. Therefore large volume of staining solution is recommended for use. FIG. 10: Structure of Coomassie Blue
11 11 H. Image analysis on the gel After the gel has been destained the objective is to quantify the mass of the proteins from the instensity of the dye that is stuck to it. An exact quantification has its own complications and I shall explain how far one can go in this goal. Basically the idea is to see how the intensity of the gel varies as a function of the protein mass and this requires a way of quantifying the intensity. For this the gel is photographed and the sum of the pixel values is taken as a proxy for the intensity since one knows that bighter points in the picture have higher pixel values. Then all analysis can be done using this set of numbers as obtained by the following process of digitizing the gel. The exact steps in the process are as follows, The last destaining solution is not transferred out of the tray. A pair of soft smooth transparent plastic sheets is cut out of the size little bigger than the gel and these are joint at one edge. Usually this is made by cutting out from the edge of plastic packets. Keeping the gel dipped inside the solution a hard plastic is slid underneath it and the gel is lifted out of the solution on it. This method ensures that no air bubbles are trapped between the gel and the plastic. Then gently the gel is slipped on to one of the pair of soft palstic sheets from the hard plastic by sliding it onto it keeping the leading edge always in touch with a thin film of water. This ensures that no air bubbles get trapped in this stage. The second plastic film is covered over it in a similar way ensuring that no air bubbles are trapped. FIG. 11: The gel as it looks just after taking out of the destaining process This setup of the gel trapped between pairs of soft tranparent plastic sheets is gently put on a scanner and the image is taken at a resolution of 600dpi. From the image the relevant part is cut out (generally a thin rectangular strip corresponding to the dilution series) and rotated if necessary. FIG. 12: The strip of the scanned image of the gel along the dilution series
12 12 The software ImageJ is used to invert the contrast of the image and convert this inverted picture into a matrix of numbers which gets saves as a text file. FIG. 13: Inverted image of the strip of the scanned image of the gel along the dilution series Inversion is done to make the numerical analysis more intuitive so that places in the image which are brighter because of the dye attaching to the protein have higher numerical values. By looking at this picture the rectangular regions are selected in the strip which correspond to the protein spots and a C++ program was written which takes this text file as input, stores it as a matrix and sums the numbers in every rectangular region corresponding to each protein spot. I. The data analysis So at the end of the data extraction one has a set of pairs of numbers {m i, I i } where I i is the intensity of of the spot corresponding to m i mass of the protein and m i+1 = 1 2 m i. Using Mathematica7 the following two graphs are plotted, I i vs m i I i vs i The graph I i vs m i shows somewhat of a linear behaviour if the extreme values are dropped. At the higher mass end (i.e > 6µg) the intesity seems to stall and at the very low mass (i.e < 0.75µg) the intensity seems to drop sharply most likely because of being very close to the lowest detection limits of this staining procedure. At a protein mass of about 0.375µg the stain is just barely visible. A typical running of a dilution gel has 6 dilutions as shown in the photographs of the strips and the intensities as defined earlier as the sum of the matrix entries in the sub-rectangles look like, { , , , , , } corresponding to {12µg, 6µg, 3µg, 1.5µg, 0.75µg, 0.375µg}. This data can be analyzed in the following ways, One can plot the points {(1, ), (2, ), (3, ), (4, ), (5, ), (6, )} and fit the function I = A x n to it and find the optimum value of A and n for that. The way to think of this graph is that given the set of points {(m i, I i ) i {1, 2,..., 6}} one is constructing from it a set {(i, I i ) i {1, 2,..., 6}} such that i = 1 + log 2 ( m 1 m i ). Here which is m 1 and which is m 2 etc is unambiguous since we know precisely in the making of the dilution series that m i+1 = 1 2 m i. So m 1 is always the highest mass sent through the gel.
13 13 FIG. 14: A pot of the points (1 + log 2 m1 m i Mathematica7 as A and n , I i) and fitted to it is the function I = A x n with the best fit values given by FIG. 15: A plot of the points (m iµg, I i) for the same rectangular strip from the same run with BSA as above FIG. 16: A plot of the points (m iµg, I i) for another run of a dilution gel with BSA which showed the typical problems of the intensities stalling for larger masses and showing sharp drops for lower masses. These are issues with the sensitivity of the detection technique. For a fixed protein the above analysis is repeated on multiple gels by taking multiple rectangles around the stained strip and the values of intensities and the coeffcients A and n are averaged over them and the standard deviation of them is taken as a proxy for the error in their measurements. Displayed below are the values for the protein Actin. Actin that was used was extracted in-house and hence the exact amount of the concentration of the protein solution is not known. But we can still do a dilution series gel using it since not knowing the exact concentration is not a hindrance in taking half of the mass inserted into the first gel ans so on. Hence for such cases we scale the mass unit and consider the highest mass lane to have 1 unit mass Actin and the lane to the right of it has 0.5 unit mass Actin and so forth. Then the similar image analysis can be done as explained before.
14 14 Run and selection Actin Gel 1, rectangle Actin Gel 1, rectangle Actin Gel 2, rectangle Actin Gel 2, rectangle Mean Standard Deviation TABLE I: Intensities and the averages and the standard deviations for the same scaled masses of Actin in various trials Fitting the function A x on the above averaged points for Actin the values gotten are A = and n n = The relevant dependencies can be seen in the graphs below. FIG. 17: The above plot shows the fall of the intensity with 1+ logarithm of the fracional mass along with an estimate of the errors in the intensities and the curve I = is fitted to that. x
15 15 FIG. 18: The above graph shows the intensities of the Coomassie stains as a function of the scaled protein mass in arbitrary units along with an estimate of the errors in the intensities. This shows the problems of stalling at higher masses and a sharp drop of the sensitivity at the lower ends. It also shows the best-fit straight line of x that one gets on dropping the lowest mass from the data. A similar experiment as above was done with the protein BSA (Bovine Serum Albumin) and this was commercially bought. Hence in this case can know with greater confidence the amount of protein present in the sample but running of the gel gave spurious bands and hence brought in the doubt that the commercial protein isn t so pure. Further the staining data on BSA seemed to show some unnaturalness. Under the expectation that the efficency of doing the experiment increases with time only the last 2 runs of doing electrophoresis on a dilution series with the BSA are reported in the follwing table and taking two rectangles from each gel as for Actin. Run and selection 12µg 6µg 3µg 1.5µg 0.75µg 0.375µg BSA Gel 1, rectangle BSA Gel 1, rectangle BSA Gel 2, rectangle BSA Gel 2, rectangle Mean Standard Deviation TABLE II: Intensities and the averages and the standard deviations for the same masses of BSA in various trials Fitting the function A x n n = on the above averaged points for BSA the values gotten are A = and
16 16 FIG. 19: The above plot shows the fall of the intensity with 1+ logarithm of the fracional mass along with an estimate of the errors in the intensities and the curve I = is fitted to that. x FIG. 20: The above graph shows the intensities of the Coomassie stains as a function of the mass of the protein in micrograms in the BSA sample along with an estimate of the errors in the intensities. This shows the problems of stalling at higher masses and sharp drop of the sensitivity at the lower ends. It also shows the best-fit straight line of m that one gets on dropping the lowest mass from the data. J. Using the results of the analysis If the largest mass of the protein used in the dilution gel is m 0 (with respect to which the other masses are scaled) then the result of the above calculation states that there exists constants A and n such that the intensity (I) of a stain goes with mass of the protein (m) as, A I = [ ( m0 ) ] n log2 m + 1 But one must be cautious of the interpretation of this result as it stands now. One should take note that the constants A and n are NOT intrinsic to the protein but depend on m 0 as can be easily checked by doing the fit on the same set of data with the current highest mass removed and scaling everything with respect to the current second highest mass. But obviously for the same m 0, A and n will be different for different proteins.
17 But for a fixed protein if m 0 and its corresponding A and n are known then one can treat the result as a standardization such that it can now be used to predict masses of other samples of the same protein from their staining intensities. But the prediction can have two different forms as follows, Like for standardization with BSA where m 0 is known, given an unknown sample of stain intensity I its mass can be predicted to be m = 2 m 0 1 A n I 1 1 Like for standardization with Actin where m 0 is not known, given two samples of proteins of stain intensities I 1 and I 2 one can predict the ratio of the masses of the proteins as, 17 m 1 m 2 = 2 A I 1 1 n 1 A n I 2 K. Some findings/observations of my own I started filtering the destaining solution twice to ensure greater efficieny of the chemical on reuse. I devised this method of transferring the gel on the scanner by using plastic supports while keeping it always submeged in the destaining solution. This practically eliminates the risk of air bubbles getting trapped. The image analysis technique described here is of my own and I have not found that in any reference. I wondered if the staining/destaining step could be avoided by sending into the gel, protein mixed with Coomassie Blue. But then this does not work as I found out that Coomassie Blue happens to have higher mobility than the proteins and hence inside the gel it moves faster ahead without causing any staining effect. III. SOME THEORETICAL BACKGROUND The entire physics and chemistry of the process of electrophoresis is a very complicated mix of phenomenon and much of the numbers like the amount of each chemical to be used is very hard to rationalize by pure theoretical calculation and are obtained by experimental optimization. A. A basic overview of the idea One of the key ideas behind electrophoresis is that if 2 solutions of different ph and different densities containing charged molecules of different mobility can be stabilized against convective instabilities then the boundary between the two fluids can be robustly maintained in an electric field. In that case the two fluids can move maintaining the boundary or if the boundary is somehow made rigid (like through polymerization) then the charged ions can flow through it in a predictable sequence without disturbing the boundary. This predictability of the sequence gives the quite essential stacking effect in gel electrophoresis which holds the key to identification of the different protein components in the initial solution. The convective instabilities are generally avoided by either having the two fluids to be immiscible and of different densities or by polymerization like here so that convection is prohibited simply due to structural rigidity. If the boundary is moveable then the crucial observation is that inspite of the difference in mobilities of the mobile ions on the two sides the velocities at which they move have to be the same since otherwise a gap gets created at the interface through which electricity cannot conduct. Since in most situations the electric field and the mobilities are given constants the formation of the gap is prevented by the rigid polymer matrix, the system attains this dynamic quilibrium by dissociating the correct number of ions on either side. Eventually it wll be seen that whether the system can dissociate to this optimal extent depends on whether the experimentalist has chosen the right dissociation reactions and ph. And much of these optimal values have been established over years of standardization of the experiment and only some order of magnitude esimates can be gotten from pure theoretical arguments.
18 The key equation behind this is called the Kohlrausch Regulating Function which will be explored to some extent in the next section. The point to remember is that the following analysis ignores the effect of the gel and considers only a solution of ions of different mobilities in the presence of a constant potential difference. 18 B. Kohlrausch Regulating Function Some basic equations and definitions that is needed to make the above concept precise are as follows, If a charged ion moves a distance d in time t in an electric field E then one defines mobility = µ = d te If a solution contains many species of mobile ions labelled by i each of concentration [i] and mobility µ i and charge ez i where e is the charge of an electron, then the conductivity σ of the solution is, σ = e i [i]µ i z i If x i is the dissociation fraction of the species i compared to its undissociated form then the average velocity of migration of this species is given as, v i = Eµ i x i For the current purpose one can restrict to dilute solutions where one can use the definition of ph as ph = log([h + ]) where [H + ] is the numerical value of the concentration of [H + ] measured in molarity. For the dilute solutions used in electrophoresis one can keep approximating the activity of the ions by the numerical values of their concentrations expressed in molarity. For this purpose the same shall be done with regard to other similar quantities like pk a, pk b and pk w. In the context of the electrophoresis set-up that is used in this laboratory the important features that need to be kept in view are the following, There is a gel on the top which has a ph of 6.8. There is a gel at the bottom which has a ph of 8.8. There are Cl 1 ions in both the layers of the gel and in the conducting buffer. Glycine ions are present only in the buffer. The mobilities of the relevant ions in increasing order of mobilities is glycine < proteins < Cl 1 Let the ions in the upper layer be labelled as u i and the ones in the lower layers are l i. Let the mobility of u i be µ iu and have a charge z iu. Let the electric field in the upper gel be E u and the field in the lower gel be E l. Let the cross-sectional area of the gels through which the current flows be A and since the two gels are in series the current through them is the same and hence equating the currents in the two layers using the definition of conductivity given earlier one has, E u Ae i [u i ]µ iu z iu = E l Ae i [l i ]µ il z il which can be re-written as,
19 19 E l i [u i]µ iu z iu = E u i [l i]µ il z il Earlier it was argued that the velocities of the ions in the two gels have to be the same because of continuity and hence here one uses that for the upper ion whose parameters are subscripted with u and for the lower ions whose corresponding parameters are subscripted with l. Then one has from the velocity equality, Substituting this back in the earlier equation we have, E u µ u x u = E l µ l x l µ u x u i [u i]µ iu z iu = µ l x l i [l i]µ il z il The above is known as the Kohlrausch Regulating Function One of ways to see what this function does is to consider a situation when µ l x l > µ u x u and in that case one can see that if an upper ion finds itsel in the lower gel then it will move slower than the surroundings and will be eventually be overtakn by the boundary and it will return to its original location. Similarly if a lower ion inds itself in the upper gel it will move faster than the boundary and will soon overtake it and get back to its original point. Thus the system maintains a clear separating layer between the two regions. Let µ p and x p be the mobility and the dissociation fraction of the proteins and µ g and x g be for the glycine. From the experimental set-up one knows that glycines will be in the upper layer and the proteins will be in the lower layer and the chloride ions are common to both the layers but has different concentrations and hence Kohlrausch Regulating function for this case says that, µ g x g [g]µ g z g + [cu]µ c z c = µ p x p [p]µ p z p + [cl]µ c z c (using c to denote the chloride ions) By microscopic charge neutrality one has [g]z g = [cu]z c and [p]z p = [cl]z c. Using that the crucial equation that emerges is that, ( cg ( µg ) ) ( ) x g z g µp µ ( ) = ( ) c cp µp µ g µ c x p z p If the effects of the gel are not included then the above equation explains why a system of ions of different mobilities should maintain distinct separating boundaries while flowing in the presence of a constant potential difference. The analysis of the gel on this above effect is a further comlicated phenomenon which in someway can be thought of as providing a viscous drag force which imepedes the motion. But the modelling of that effect runs into complications ofthe fact that denatured protein molecules in the system tend to be cylindrical in shape and hence the direction of their approach becomes a crucial factor after a certain molecular mass number. This is what is dubbed as Non- Newtonian viscosity. Further also needs to model the effect that the static ph boundary in the gel does not disrupt the moving ph boundary of the ion solution. C. Some comment about the polymerization It is a still harder problem to model how doe the pore size of the gel vary as a function of the concentration of the polymer. As a minimal model consider that the gel consists of n n n cubical cells for evey unit cube in it and network be formed of polymer chains of unit length. Then one can approximate the pore diameter by the length of a side of the cell (say r) and then one can see that r = 1 n ( n) d and the number of polymer chains in each such cube is 3(n + 1) 2. Then simple calculation gives that if the concentration of the polymer is increased by J then number of squares per edge would becme (n + 1) J 1 and hence from that one can calculate by how much the r changes with J.
20 20 IV. CONCLUSION This experiment and the associated analysis shows a careful analysis of the limitations of being able predict mass of a protein in a given sample by looking at intensitities of dye stains. In the process the use of running a serial dilution of the protein solution through an SDS-PAGE electrophoresis set-up is found to be helpful to find out the dependence of the intensity of the stain on the mass. Analysis of that shows that though intensities of stain seem to show not so regular behaviour with the mass of the protein the intensities show a power-law fall with masses of the protein with exponents and pre-factors which seem robust across experiments. Hence as an end result one can get standardization curves which can be used to either predict masses of proteins in a random solution if in one sample the mass is exactly known or ratios of masses in different samples in any case. Doing of the experiment also introduced me to new experimental skills involved in handling microliters of fluids and doing image analysis. V. ACKNOWLEDGEMENT I would like to express my deep gratitude for Prof.Roop Mallick for letting me intern in his laboratory for these 2 months and for giving me immense freedom to explore various possibilities and suggesting crucial checks and cautions while analyzing the results. I was fortunate to receive and extraordinary amount of help from Pradeep. It was heartening to see him even compromise on his examination peparations and his own research to help me with every experiment that I tried and especially for patiently teaching me all the basic techniques. For every problem in the laboratory I found Pradeep s kind help to save the day. Further I would like to thank Ashim for helping me through a Western Blotting experiment and also for his timely and sharp insights with feasibility of experiments. I would like to thank Ravi for letting me use some of the solutions of Actin protein which he along with the others had painstakingly extracted and purified. I would like to thank members of Prof.Ulhas Kolthur s lab for letting me use their water heater and antibodies ever so often. Once I also had to use the water bath in Prof.Shubha Tole s lab. Thanks are also due to Prof. Gotam. K. Jarori s lab for letting me use their 30% Acrylamide solutions. Lastly I would like to thank the Suject Board of Physics at TIFR for providing this enriching experience. 5 Electrobloting of proteins from polyacrylamide gels by Michael.J.Dunn, published in Protein Purification Protocols edited by Paul Cutler 5 Using Antibodies by Ed Harlow and David Lane 5 Disc Electrophoresis-I by Leonard Ornstein 5 Isotachophoresis-Some fundamental aspects by Jozef Leonardus Beckers 5 Wikipedia
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis):
 SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
SDS-polyacrylamide gel electrophoresis
 SDS-polyacrylamide gel electrophoresis Protein Isolation and Purification Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells,
SDS-polyacrylamide gel electrophoresis Protein Isolation and Purification Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells,
Protocol for 2D-E. Protein Extraction
 Protocol for 2D-E Protein Extraction Reagent 1 inside the ReadyPrep TM Sequential Extraction kit (in powder form) 50ml of deionized water is used to dissolve all the Reagent 1. The solution is known as
Protocol for 2D-E Protein Extraction Reagent 1 inside the ReadyPrep TM Sequential Extraction kit (in powder form) 50ml of deionized water is used to dissolve all the Reagent 1. The solution is known as
Protein separation and characterization
 Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: tech@aladdin-e.com Email EU: eutech@aladdin-e.com Email Asia Pacific: cntech@aladdin-e.com Protein separation
Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: tech@aladdin-e.com Email EU: eutech@aladdin-e.com Email Asia Pacific: cntech@aladdin-e.com Protein separation
Protease Inhibitor Cocktail A (1 tablet / 7 10 ml, Roche Cat# ) Protease inhibitor Cocktail B (0.5ml per 250ml, Calbiochem Cat# )
 Protocol for Western Blotting Tissue/Cell Sample Preparation Lysis Buffer 1 (ph8.0) o 50mM Tris-Cl o 150mM NaCl o 1% v/v NP40 o protease inhibitor cocktail A/B Lysis Buffer 2 (RIPA) (ph 8.0) o 50mM Tris-Cl
Protocol for Western Blotting Tissue/Cell Sample Preparation Lysis Buffer 1 (ph8.0) o 50mM Tris-Cl o 150mM NaCl o 1% v/v NP40 o protease inhibitor cocktail A/B Lysis Buffer 2 (RIPA) (ph 8.0) o 50mM Tris-Cl
Student Guide. bluegel TM Electrophoresis Rainbow Lab. 1. Synopsis
 bluegel TM Electrophoresis Rainbow Lab Student Guide Contents 1. Synopsis p. 1 2. Background p. 2 3. Student laboratory guide p. 7 4. Student data table p. 13 5. Study questions p. 14 1. Synopsis In this
bluegel TM Electrophoresis Rainbow Lab Student Guide Contents 1. Synopsis p. 1 2. Background p. 2 3. Student laboratory guide p. 7 4. Student data table p. 13 5. Study questions p. 14 1. Synopsis In this
Chromatography Lab # 4
 Chromatography Lab # 4 Chromatography is a method for separating mixtures based on differences in the speed at which they migrate over or through a stationary phase which means that a complex mixture will
Chromatography Lab # 4 Chromatography is a method for separating mixtures based on differences in the speed at which they migrate over or through a stationary phase which means that a complex mixture will
Module 3. Establishing Standard Curve (Bradford Assay)
 Module 3. Establishing Standard Curve (Bradford Assay) 1. Melissa Myoglobin and the Missing Molecules Melissa Myoglobin had a little tube filled with a clear protein solution. To do more research with
Module 3. Establishing Standard Curve (Bradford Assay) 1. Melissa Myoglobin and the Missing Molecules Melissa Myoglobin had a little tube filled with a clear protein solution. To do more research with
E lectrophores is with S DS buffer kit ph 8.0 INS T R UC T IONS
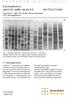 A M E R S H A M B I O S C I E N C E S E lectrophores is with S DS buffer kit ph 8.0 INS T R UC T IONS C leang elª with S DS B uffer K it for horizontal S DS electrophores is Ð Fig. 1. SD S electrophoresis
A M E R S H A M B I O S C I E N C E S E lectrophores is with S DS buffer kit ph 8.0 INS T R UC T IONS C leang elª with S DS B uffer K it for horizontal S DS electrophores is Ð Fig. 1. SD S electrophoresis
Molecular Rainbow Dye Electrophoresis Student Materials
 Dye Electrophoresis Student Materials Introduction. 2 Pre-Lab Questions 4 Lab Protocol. 5 Data Collection Worksheet 6 Post-Lab Questions and Analysis.. 7 Last updated: 10/23/2017 Introduction Take just
Dye Electrophoresis Student Materials Introduction. 2 Pre-Lab Questions 4 Lab Protocol. 5 Data Collection Worksheet 6 Post-Lab Questions and Analysis.. 7 Last updated: 10/23/2017 Introduction Take just
DNA can be extracted from the following sample types using this procedure: Archived
 Sample types Principle Safety Equipment and supplies DNA can be extracted from the following sample types using this procedure: concentrated DNA samples (e.g., blood, saliva, non-contact samples) hair
Sample types Principle Safety Equipment and supplies DNA can be extracted from the following sample types using this procedure: concentrated DNA samples (e.g., blood, saliva, non-contact samples) hair
Principles of Thin Layer Chromatography
 REVISED & UPDATED Edvo-Kit #113 Principles of Thin Layer Chromatography Experiment Objective: The objective of this experiment is to gain an understanding of the theory and methods of thin layer chromatography.
REVISED & UPDATED Edvo-Kit #113 Principles of Thin Layer Chromatography Experiment Objective: The objective of this experiment is to gain an understanding of the theory and methods of thin layer chromatography.
SDS-PAGE of Proteins. Protocol INTRODUCTION MATERIALS. Please cite as: CSH Protocols; 2006; doi: /pdb.prot4313. Richard J.
 Please cite as: CSH Protocols; 2006; doi:10.1101/pdb.prot4313 Protocol SDS-PAGE of Proteins Richard J. Simpson This protocol was adapted from "Concentrating Solutions of Protein," in Appendix 5 of Purifying
Please cite as: CSH Protocols; 2006; doi:10.1101/pdb.prot4313 Protocol SDS-PAGE of Proteins Richard J. Simpson This protocol was adapted from "Concentrating Solutions of Protein," in Appendix 5 of Purifying
TWO ENZYME CATALYSIS OVERVIEW OBJECTIVES INTRODUCTION
 TWO ENZYME CATALYSS OVERVEW n this lab you will: 1. observe the conversion of hydrogen peroxide (H 2 0 2 ) to water and oxygen gas by the enzyme catalase, and 2. measure the amount of oxygen generated
TWO ENZYME CATALYSS OVERVEW n this lab you will: 1. observe the conversion of hydrogen peroxide (H 2 0 2 ) to water and oxygen gas by the enzyme catalase, and 2. measure the amount of oxygen generated
BY PRACTICAL SKILLS IN BIOLOGY
 BY1101 - PRACTICAL SKILLS IN BIOLOGY Practical 3 Molecular Techniques 3 Protein Purification by Gel-Filtration and Thurs 18 and Fri 19 October 2012 CONTINUING OUR INVESTIGATION OF CHROMATOGRAPHY Some molecular
BY1101 - PRACTICAL SKILLS IN BIOLOGY Practical 3 Molecular Techniques 3 Protein Purification by Gel-Filtration and Thurs 18 and Fri 19 October 2012 CONTINUING OUR INVESTIGATION OF CHROMATOGRAPHY Some molecular
Analytical Technologies in Biotechnology Prof. Dr. Ashwani K. Sharma Department of Biotechnology Indian Institute of Technology, Roorkee
 Analytical Technologies in Biotechnology Prof. Dr. Ashwani K. Sharma Department of Biotechnology Indian Institute of Technology, Roorkee Module - 4 Electrophoresis Lecture - 1 Basis Concept in Electrophoresis
Analytical Technologies in Biotechnology Prof. Dr. Ashwani K. Sharma Department of Biotechnology Indian Institute of Technology, Roorkee Module - 4 Electrophoresis Lecture - 1 Basis Concept in Electrophoresis
Inserting grids into capsule Removing capsules from grid box Immunolabeling using template guide Preparing for TEM insertion
 Protocol Immunolabeling protocols are easily adapted to mprep/g processing. This document illustrates a typical protocol using a primary antibody and a secondary antibody conjugated to colloidal gold,
Protocol Immunolabeling protocols are easily adapted to mprep/g processing. This document illustrates a typical protocol using a primary antibody and a secondary antibody conjugated to colloidal gold,
Module : 9 Electrophoretic Separation Methods
 Module : 9 Electrophoretic Separation Methods Dr. Sirshendu De Professor, Department of Chemical Engineering Indian Institute of Technology, Kharagpur e-mail: sde@che.iitkgp.ernet.in Keywords: Separation
Module : 9 Electrophoretic Separation Methods Dr. Sirshendu De Professor, Department of Chemical Engineering Indian Institute of Technology, Kharagpur e-mail: sde@che.iitkgp.ernet.in Keywords: Separation
A Gene Discovery Lab Manual For Undergraduates:
 A Gene Discovery Lab Manual For Undergraduates: Searching For Genes Required To Make A Seed Honors Collegium 70AL Summer 2014 By Kelli Henry Anhthu Q. Bui Brandon Le Bob Goldberg Department of Molecular,
A Gene Discovery Lab Manual For Undergraduates: Searching For Genes Required To Make A Seed Honors Collegium 70AL Summer 2014 By Kelli Henry Anhthu Q. Bui Brandon Le Bob Goldberg Department of Molecular,
Sero: p30 Crossover Electrophoresis for Semen
 Safety Principle Equipment and supplies SAFETY WARNING Electrophoretic procedures have electrical hazards associated with them. Therefore, at minimum, the presence of two analysts is required during these
Safety Principle Equipment and supplies SAFETY WARNING Electrophoretic procedures have electrical hazards associated with them. Therefore, at minimum, the presence of two analysts is required during these
In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE
 In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE (Lamond Lab / April 2008)! Perform all the pipetting steps in a laminar flow hood. We routinely do our digestions in our TC room hoods.
In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE (Lamond Lab / April 2008)! Perform all the pipetting steps in a laminar flow hood. We routinely do our digestions in our TC room hoods.
PROTOCOL: GCP Protein Quantification Assay and QC (QUANT)
 PROTOCOL: GCP Protein Quantification Assay and QC (QUANT) Purpose To determine overall protein yield and histone purity after histone extraction. SDS PAGE gels are not required, only when changing variables
PROTOCOL: GCP Protein Quantification Assay and QC (QUANT) Purpose To determine overall protein yield and histone purity after histone extraction. SDS PAGE gels are not required, only when changing variables
THE DYE/INDICATOR LAB SEPARATION OF MOLECULES USING AGAROSE GEL ELECTROPHORESIS
 THE DYE/INDICATOR LAB SEPARATION OF MOLECULES USING AGAROSE GEL ELECTROPHORESIS OBJECTIVES: YOU WILL BE ABLE TO PERFORM THE FOLLOWING ACTIVITIES. Make and load the wells of an agarose gel. Electrophorese
THE DYE/INDICATOR LAB SEPARATION OF MOLECULES USING AGAROSE GEL ELECTROPHORESIS OBJECTIVES: YOU WILL BE ABLE TO PERFORM THE FOLLOWING ACTIVITIES. Make and load the wells of an agarose gel. Electrophorese
user manual blotting Hoefer PR648 Slot blot manifold um PR648-IM/Rev.F0/08-12
 user manual blotting Hoefer PR648 Slot blot manifold um PR648-IM/Rev.F0/08-12 Page finder Introduction...1 Specifications...2 Procedure for standard use of the Hoefer PR648...3 Setting up the Hoefer PR648...3
user manual blotting Hoefer PR648 Slot blot manifold um PR648-IM/Rev.F0/08-12 Page finder Introduction...1 Specifications...2 Procedure for standard use of the Hoefer PR648...3 Setting up the Hoefer PR648...3
EXPERIMENT 15. USING CONDUCTIVITY TO LOOK AT SOLUTIONS: DO WE HAVE CHARGED IONS OR NEUTRAL MOLECULES? rev 7/09
 EXPERIMENT 15 USING CONDUCTIVITY TO LOOK AT SOLUTIONS: DO WE AVE CARGED IONS OR NEUTRAL MOLECULES? rev 7/09 GOAL After you complete this experiment, you should have a better understanding of aqueous solutions
EXPERIMENT 15 USING CONDUCTIVITY TO LOOK AT SOLUTIONS: DO WE AVE CARGED IONS OR NEUTRAL MOLECULES? rev 7/09 GOAL After you complete this experiment, you should have a better understanding of aqueous solutions
Biochemistry. Biochemical Techniques. 01 Electrophoresis : Basic Concepts
 Description of Module Subject Name Paper Name 12 Module Name/Title 01 Electrophoresis: Basic Concept 1. Objectives 1.1 To understand basic concept of electrophoresis 1.2 To explain what determines charge
Description of Module Subject Name Paper Name 12 Module Name/Title 01 Electrophoresis: Basic Concept 1. Objectives 1.1 To understand basic concept of electrophoresis 1.2 To explain what determines charge
Protein extraction and quantification by Teresa Fan, University of Kentucky BUFFER PREPARATION AND STORAGE
 Protein extraction and quantification by Teresa Fan, University of Kentucky Note: This procedure follows [Fan_Extract_Polar_Lipid_Prot_SOP] Step 11 (middle part, in 1.5 ml tube). BUFFER PREPARATION AND
Protein extraction and quantification by Teresa Fan, University of Kentucky Note: This procedure follows [Fan_Extract_Polar_Lipid_Prot_SOP] Step 11 (middle part, in 1.5 ml tube). BUFFER PREPARATION AND
Biological Sciences 11 Spring Experiment 4. Protein crosslinking
 Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Electrolysis: Splitting Water Student Advanced Version
 Electrolysis: Splitting Water Student Advanced Version In this lab you will use a battery to perform electrolysis, or chemical decomposition, of different aqueous solutions (like water) to produce gases
Electrolysis: Splitting Water Student Advanced Version In this lab you will use a battery to perform electrolysis, or chemical decomposition, of different aqueous solutions (like water) to produce gases
LAB 6 - GRAVITATIONAL AND PASSIVE FORCES
 83 Name Date Partners LAB 6 - GRAVITATIONAL AND PASSIVE FORCES OBJECTIVES OVERVIEW And thus Nature will be very conformable to herself and very simple, performing all the great Motions of the heavenly
83 Name Date Partners LAB 6 - GRAVITATIONAL AND PASSIVE FORCES OBJECTIVES OVERVIEW And thus Nature will be very conformable to herself and very simple, performing all the great Motions of the heavenly
Ka Acid Dissociation Constant Minneapolis Community and Technical College Principles of Chemistry II, C1152 v.1.16
 Ka Acid Dissociation Constant Minneapolis Community and Technical College Principles of Chemistry II, C1152 v.1.16 I. Introduction Monoprotic acetic acid, CH 3 COOH is sometimes written as HCH 3 COO, HC
Ka Acid Dissociation Constant Minneapolis Community and Technical College Principles of Chemistry II, C1152 v.1.16 I. Introduction Monoprotic acetic acid, CH 3 COOH is sometimes written as HCH 3 COO, HC
Recommended Procedures for Labeling. Labeling Proteins with Amine-Reactive ATTO-Labels (NHS-Esters) Introduction
 Recommended Procedures for Labeling Introduction ATTO-TEC offers a large variety of high-quality dyes for labeling amino and thiol groups. ATTO reactive dyes cover the spectral region from 350 nm in the
Recommended Procedures for Labeling Introduction ATTO-TEC offers a large variety of high-quality dyes for labeling amino and thiol groups. ATTO reactive dyes cover the spectral region from 350 nm in the
ph Measurement and its Applications
 ph Measurement and its Applications Objectives: To measure the ph of various solutions using indicators and ph meters. To perform a ph titration. To create and study buffer solutions. To determine the
ph Measurement and its Applications Objectives: To measure the ph of various solutions using indicators and ph meters. To perform a ph titration. To create and study buffer solutions. To determine the
The effects of histone and glycosaminoglycans on human factor Xa and antithrombin III interactions
 Bowling Green State University ScholarWorks@BGSU Honors Projects Honors College Fall 12-12-2013 The effects of histone and glycosaminoglycans on human factor and antithrombin III interactions Amy Thomas
Bowling Green State University ScholarWorks@BGSU Honors Projects Honors College Fall 12-12-2013 The effects of histone and glycosaminoglycans on human factor and antithrombin III interactions Amy Thomas
Soil Cation Analysis Using High-Performance Capillary Zone Electrophoresis Last Modified: October 20, 2006
 Soil Cation Analysis Using High-Performance Capillary Zone Electrophoresis Last Modified: October 20, 2006 Introduction: Capillary electrophoresis (CE) is a relatively new, but rapidly growing separation
Soil Cation Analysis Using High-Performance Capillary Zone Electrophoresis Last Modified: October 20, 2006 Introduction: Capillary electrophoresis (CE) is a relatively new, but rapidly growing separation
Movement of Molecules Biology Concepts of Biology 3.1
 Movement of Molecules Biology 100 - Concepts of Biology 3.1 Name Instructor Lab Section Objectives: To gain an understanding of: The basic principles of osmosis and diffusion Brownian motion The effects
Movement of Molecules Biology 100 - Concepts of Biology 3.1 Name Instructor Lab Section Objectives: To gain an understanding of: The basic principles of osmosis and diffusion Brownian motion The effects
Acid-Base Titration. Sample
 Acid-Base Titration Computer 7 A titration is a process used to determine the volume of a solution that is needed to react with a given amount of another substance. In this experiment, your goal is to
Acid-Base Titration Computer 7 A titration is a process used to determine the volume of a solution that is needed to react with a given amount of another substance. In this experiment, your goal is to
EXPERIMENT 9 ENTHALPY OF REACTION HESS S LAW
 EXPERIMENT 9 ENTHALPY OF REACTION HESS S LAW INTRODUCTION Chemical changes are generally accompanied by energy changes; energy is absorbed or evolved, usually as heat. Breaking chemical bonds in reactants
EXPERIMENT 9 ENTHALPY OF REACTION HESS S LAW INTRODUCTION Chemical changes are generally accompanied by energy changes; energy is absorbed or evolved, usually as heat. Breaking chemical bonds in reactants
Supernatant: The liquid layer lying above the solid layer after a precipitation reaction occurs.
 Limiting Reagent Introduction The quantities of substances involved in a chemical reaction represented by a balanced equation are often referred to as stoichiometric amounts. Solution stoichiometry is
Limiting Reagent Introduction The quantities of substances involved in a chemical reaction represented by a balanced equation are often referred to as stoichiometric amounts. Solution stoichiometry is
Polymer Chemistry Prof. Dibakar Dhara Department of Chemistry Indian Institute of Technology, Kharagpur
 Polymer Chemistry Prof. Dibakar Dhara Department of Chemistry Indian Institute of Technology, Kharagpur Lecture - 10 Radical Chain Polymerization (Contd.) (Refer Slide Time: 00:28) Welcome back, and we
Polymer Chemistry Prof. Dibakar Dhara Department of Chemistry Indian Institute of Technology, Kharagpur Lecture - 10 Radical Chain Polymerization (Contd.) (Refer Slide Time: 00:28) Welcome back, and we
Enzyme Catalysis Lab
 AP Biology Name: Enzyme Catalysis Lab Objectives In this laboratory, you will observe the role of an enzyme (catalase) in conversion of hydrogen peroxide (H 2 O 2 ) to water and oxygen determine the rate
AP Biology Name: Enzyme Catalysis Lab Objectives In this laboratory, you will observe the role of an enzyme (catalase) in conversion of hydrogen peroxide (H 2 O 2 ) to water and oxygen determine the rate
8.6 Copper Plating & Electrolysis. Grade 8 Activity Plan
 8.6 Copper Plating & Electrolysis Grade 8 Activity Plan 1 Reviews and Updates 2 8.6 Copper Plating & Electrolysis Objectives: 1. To understand how the laws of attraction govern the formation of ions and
8.6 Copper Plating & Electrolysis Grade 8 Activity Plan 1 Reviews and Updates 2 8.6 Copper Plating & Electrolysis Objectives: 1. To understand how the laws of attraction govern the formation of ions and
Name SCS- Date. Experiment 5: Hydrogen Formation and Reaction with Oxygen
 Name SCS- Date Experiment 5: Hydrogen Formation and Reaction with Oxygen Aims: What are the energy changes associated with chemical bond formation and breakage? How are changes in potential energy related
Name SCS- Date Experiment 5: Hydrogen Formation and Reaction with Oxygen Aims: What are the energy changes associated with chemical bond formation and breakage? How are changes in potential energy related
Station 1 Water is a polar molecule and has a very unique structure
 Station 1 Water is a polar molecule and has a very unique structure A water molecule, because of its shape, is a polar molecule. That is, it has one side that is positively charged and one side that is
Station 1 Water is a polar molecule and has a very unique structure A water molecule, because of its shape, is a polar molecule. That is, it has one side that is positively charged and one side that is
Chemical Reactions: The Copper Cycle
 1 Chemical Reactions: The Copper Cycle ORGANIZATION Mode: pairs assigned by instructor Grading: lab notes, lab performance and post-lab report Safety: Goggles, closed-toe shoes, lab coat, long pants/skirts
1 Chemical Reactions: The Copper Cycle ORGANIZATION Mode: pairs assigned by instructor Grading: lab notes, lab performance and post-lab report Safety: Goggles, closed-toe shoes, lab coat, long pants/skirts
Peroxidase Enzyme Lab
 Peroxidase Enzyme Lab An enzyme is a type of protein known as a biological catalyst. A catalyst speeds up chemical reactions by lowering the activation energy required. Enzymes, biological catalysts carry
Peroxidase Enzyme Lab An enzyme is a type of protein known as a biological catalyst. A catalyst speeds up chemical reactions by lowering the activation energy required. Enzymes, biological catalysts carry
Exercise 2-4. Titration of a Buffer Solution EXERCISE OBJECTIVES
 Exercise 2-4 Titration of a Buffer Solution EXERCISE OBJECTIVES To define the terms buffer solution and buffer capacity; To titrate a buffer solution with a weak acid solution; To plot a graph using the
Exercise 2-4 Titration of a Buffer Solution EXERCISE OBJECTIVES To define the terms buffer solution and buffer capacity; To titrate a buffer solution with a weak acid solution; To plot a graph using the
Prealbumin (Mouse) ELISA KitI
 Prealbumin (Mouse) ELISA KitI Catalog Number KA2070 96 assays Version: 04 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of
Prealbumin (Mouse) ELISA KitI Catalog Number KA2070 96 assays Version: 04 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of
6 Acid Base Titration
 E x p e r i m e n t Acid Base Titration Experiment : http://genchemlab.wordpress.com/-titration/ objectives To understand the concept of titration. To explain the difference between the analyte and standard
E x p e r i m e n t Acid Base Titration Experiment : http://genchemlab.wordpress.com/-titration/ objectives To understand the concept of titration. To explain the difference between the analyte and standard
INTRODUCTION. Amino acids occurring in nature have the general structure shown below:
 Biochemistry I Laboratory Amino Acid Thin Layer Chromatography INTRODUCTION The primary importance of amino acids in cell structure and metabolism lies in the fact that they serve as building blocks for
Biochemistry I Laboratory Amino Acid Thin Layer Chromatography INTRODUCTION The primary importance of amino acids in cell structure and metabolism lies in the fact that they serve as building blocks for
Protein Quantification Kit (Bradford Assay)
 Protein Quantification Kit (Bradford Assay) Booklet Item NO. KTD3002 Product Name Protein Quantification Kit (Bradford Assay) ATTENTION For laboratory research use only. Not for clinical or diagnostic
Protein Quantification Kit (Bradford Assay) Booklet Item NO. KTD3002 Product Name Protein Quantification Kit (Bradford Assay) ATTENTION For laboratory research use only. Not for clinical or diagnostic
Materials: Micropipettes (2-20 µl range pipette, µl range, µl range), tips, test tubes with color dye, well plates
 Virtually every chemical reaction in a lab or manufacturing facility, as in cells, occurs in a watery environment or solution. A lab technician, therefore, must be able to quickly prepare any volume of
Virtually every chemical reaction in a lab or manufacturing facility, as in cells, occurs in a watery environment or solution. A lab technician, therefore, must be able to quickly prepare any volume of
THE CATHOLIC UNIVERSITY OF EASTERN AFRICA A. M. E. C. E. A
 THE CATHOLIC UNIVERSITY OF EASTERN AFRICA A. M. E. C. E. A MAIN EXAMINATION P.O. Box 62157 00200 Nairobi - KENYA Telephone: 891601-6 Fax: 254-20-891084 E-mail:academics@cuea.edu JANUARY APRIL 2014 TRIMESTER
THE CATHOLIC UNIVERSITY OF EASTERN AFRICA A. M. E. C. E. A MAIN EXAMINATION P.O. Box 62157 00200 Nairobi - KENYA Telephone: 891601-6 Fax: 254-20-891084 E-mail:academics@cuea.edu JANUARY APRIL 2014 TRIMESTER
OESTREICH LAB CHIP PROTOCOL
 OESTREICH LAB CHIP PROTOCOL Buffers (all starred (*) buffers must be autoclaved before use) 10X Cell Lysis Buffer*: 100mL 0.214g Potassium Acetate (KOAc) = 10mM 0.147g Magnesium Acetate (MgAc) = 15mM 10mL
OESTREICH LAB CHIP PROTOCOL Buffers (all starred (*) buffers must be autoclaved before use) 10X Cell Lysis Buffer*: 100mL 0.214g Potassium Acetate (KOAc) = 10mM 0.147g Magnesium Acetate (MgAc) = 15mM 10mL
EXPERIMENT: LIMITING REAGENT. NOTE: Students should have moles of reactants in DATASHEET converted into masses in grams prior to the lab period.
 Revised 12/2015 EXPERIMENT: LIMITING REAGENT Chem 1104 Lab NOTE: Students should have moles of reactants in DATASHEET converted into masses in grams prior to the lab period. INTRODUCTION Limiting reactant
Revised 12/2015 EXPERIMENT: LIMITING REAGENT Chem 1104 Lab NOTE: Students should have moles of reactants in DATASHEET converted into masses in grams prior to the lab period. INTRODUCTION Limiting reactant
PRACTICAL 3 ph AND BUFFERS
 PRACTICAL 3 ph AND BUFFERS ph and Buffers Structure 3.1 Introduction 3.2 ph and Buffers: Basic Concept 3.2.1 ph 3.2.2 Buffers and Buffer Solutions 3.3 Methods for Determining ph Experiment 1: Measurement
PRACTICAL 3 ph AND BUFFERS ph and Buffers Structure 3.1 Introduction 3.2 ph and Buffers: Basic Concept 3.2.1 ph 3.2.2 Buffers and Buffer Solutions 3.3 Methods for Determining ph Experiment 1: Measurement
Flushing Out the Moles in Lab: The Reaction of Calcium Chloride with Carbonate Salts
 Flushing Out the Moles in Lab: The Reaction of Calcium Chloride with Carbonate Salts Pre-lab Assignment: Reading: 1. Chapter sections 3.3, 3.4, 3.7 and 4.2 in your course text. 2. This lab handout. Questions:
Flushing Out the Moles in Lab: The Reaction of Calcium Chloride with Carbonate Salts Pre-lab Assignment: Reading: 1. Chapter sections 3.3, 3.4, 3.7 and 4.2 in your course text. 2. This lab handout. Questions:
HBeAg and HBeAg Ab ELISA Kit
 HBeAg and HBeAg Ab ELISA Kit Cat. No.:DEIA3532 Pkg.Size:96T Intended use This kit is designed for detection of HBeAg and HBeAg Ab. General Description The solid phase of HBEAG AND HBEAG ANTIBODY ELISA
HBeAg and HBeAg Ab ELISA Kit Cat. No.:DEIA3532 Pkg.Size:96T Intended use This kit is designed for detection of HBeAg and HBeAg Ab. General Description The solid phase of HBEAG AND HBEAG ANTIBODY ELISA
Functional Genomics Research Stream. Research Meeting: February 7, 2012 Reagent Production, Buffers & ph
 Functional Genomics Research Stream Research Meeting: February 7, 2012 Reagent Production, Buffers & ph Section VII Yeast Growth Flow Wednesday Thurs. morning S288C S288C colony dilute to ~0.2 S288C other
Functional Genomics Research Stream Research Meeting: February 7, 2012 Reagent Production, Buffers & ph Section VII Yeast Growth Flow Wednesday Thurs. morning S288C S288C colony dilute to ~0.2 S288C other
Sampling and DNA Extraction from Wastewater Activated Sludge Standard Protocol
 Sampling and DNA Extraction from Wastewater Activated Sludge Standard Protocol Version 7.0 Skill Prerequisites: DNA handling Introduction This protocol explains sampling and DNA extraction from activated
Sampling and DNA Extraction from Wastewater Activated Sludge Standard Protocol Version 7.0 Skill Prerequisites: DNA handling Introduction This protocol explains sampling and DNA extraction from activated
Photosynthesis. Introduction: Objectives:
 Photosynthesis Introduction: Photosynthesis is a process in which plants convert light energy (sunlight) into usable chemical energy (carbohydrates). Photosynthesis involves two simultaneous processes:
Photosynthesis Introduction: Photosynthesis is a process in which plants convert light energy (sunlight) into usable chemical energy (carbohydrates). Photosynthesis involves two simultaneous processes:
Bis sulfone Reagents. Figure 1.
 Bis sulfone Reagents An intact IgG molecule has four accessible inter chain disulfide bonds that can be reduced to form eight free cysteine thiols, which can serve as sites for conjugation. The reaction
Bis sulfone Reagents An intact IgG molecule has four accessible inter chain disulfide bonds that can be reduced to form eight free cysteine thiols, which can serve as sites for conjugation. The reaction
What Do You Think? Investigate GOALS
 Activity 7 Polymers GOALS In this activity you will: Make a polymer-based material that has properties different from other states of matter that you have studied. Observe the material s properties and
Activity 7 Polymers GOALS In this activity you will: Make a polymer-based material that has properties different from other states of matter that you have studied. Observe the material s properties and
Alkaline Phosphatase Labeling Kit-NH2
 Alkaline Phosphatase Labeling Kit-NH2 Catalog Number KA0001 1 Kit Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Alkaline Phosphatase Labeling Kit-NH2 Catalog Number KA0001 1 Kit Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Air Quality 2 - Determination of Cd and Pb by Anodic Stripping Voltammetry
 Air Quality 2 - Determination of Cd and Pb by Anodic Stripping Voltammetry Introduction: In the first experiment of this set, you measured the concentration of benzene in gasoline, and it is a reasonable
Air Quality 2 - Determination of Cd and Pb by Anodic Stripping Voltammetry Introduction: In the first experiment of this set, you measured the concentration of benzene in gasoline, and it is a reasonable
Instantaneous and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA Using a ph-assisted and Surfactant-Free Route
 Supporting Information Instantaneous and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA Using a ph-assisted and Surfactant-Free Route Xu Zhang,, Mark R. Servos and Juewen Liu *
Supporting Information Instantaneous and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA Using a ph-assisted and Surfactant-Free Route Xu Zhang,, Mark R. Servos and Juewen Liu *
LAB 05 Enzyme Action
 LAB 05 Enzyme Action Objectives: Name the substrate and products of the peroxidase-catalyzed reaction. To understand the terms: enzyme, activation energy, active site, ph, and denaturation. Distinguish
LAB 05 Enzyme Action Objectives: Name the substrate and products of the peroxidase-catalyzed reaction. To understand the terms: enzyme, activation energy, active site, ph, and denaturation. Distinguish
College of the Canyons Biotechnology Program
 College of the Canyons Biotechnology Program Gel Filtration: Version A Gel Filtration is a chromatographic technique where by scientists use to separate one substance from another by a process known as
College of the Canyons Biotechnology Program Gel Filtration: Version A Gel Filtration is a chromatographic technique where by scientists use to separate one substance from another by a process known as
Effect of alkaline ph on sunflower 11S protein
 J Biosci, Vol. 11, Numbers 1 4, March 1987, pp. 351 360. Printed in India Effect of alkaline ph on sunflower 11S protein G. SRIPAD* and M. S. NARASINGA RAO Protein Technology Discipline, Central Food Technological
J Biosci, Vol. 11, Numbers 1 4, March 1987, pp. 351 360. Printed in India Effect of alkaline ph on sunflower 11S protein G. SRIPAD* and M. S. NARASINGA RAO Protein Technology Discipline, Central Food Technological
Bradford Reagent, 5x
 INSTRUCTION MANUAL Bradford Reagent, 5x Reagent for protein quantification (Cat. No. 39222) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 HeidelbergPhone +49-6221-138400, Fax +49-6221-1384010 e-mail:
INSTRUCTION MANUAL Bradford Reagent, 5x Reagent for protein quantification (Cat. No. 39222) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 HeidelbergPhone +49-6221-138400, Fax +49-6221-1384010 e-mail:
OMX-S pro Instruction Manual* Oct/07
 OMX-S Instruction Manual OMX-S for rapid in-gel digestion OMX-S pro Instruction Manual* Oct/07 Part No.: 17001 Table of Contents 1. Introduction...2 2. Content and precautions...2 2.1. Content...2 2.2.
OMX-S Instruction Manual OMX-S for rapid in-gel digestion OMX-S pro Instruction Manual* Oct/07 Part No.: 17001 Table of Contents 1. Introduction...2 2. Content and precautions...2 2.1. Content...2 2.2.
Acid-Base Titration. Evaluation copy
 Acid-Base Titration Computer 7 A titration is a process used to determine the volume of a solution that is needed to react with a given amount of another substance. In this experiment, your goal is to
Acid-Base Titration Computer 7 A titration is a process used to determine the volume of a solution that is needed to react with a given amount of another substance. In this experiment, your goal is to
IRDye 800CW Protein Labeling Kit Low MW
 IRDye 800CW Protein Labeling Kit Low MW Developed for: Aerius, and Odyssey Family of Imagers Please refer to your manual to confirm that this protocol is appropriate for the applications compatible with
IRDye 800CW Protein Labeling Kit Low MW Developed for: Aerius, and Odyssey Family of Imagers Please refer to your manual to confirm that this protocol is appropriate for the applications compatible with
LABORATORY 2. ENZYME CATALYSIS
 LABORATORY 2 STUDENT GUIDE LABORATORY 2. ENZYME CATALYSIS Objectives In this laboratory, you will observe the role of an enzyme (catalase) in conversion of hydrogen peroxide (H 2 O 2 ) to water and oxygen
LABORATORY 2 STUDENT GUIDE LABORATORY 2. ENZYME CATALYSIS Objectives In this laboratory, you will observe the role of an enzyme (catalase) in conversion of hydrogen peroxide (H 2 O 2 ) to water and oxygen
PreLAD: b. KHP is a monoprotic acid, what are the number of moles of ionizable H + in the approximately 0.25 g of KHP?
 LAD G.1 (pg! 1 of 6! ) What % of vinegar is really acetic acid? Name Per What part of the solution is really HC2H3O2 aka CH3COOH? What is the Ka of acetic acid? Introduction: The acid in vinegar is acetic
LAD G.1 (pg! 1 of 6! ) What % of vinegar is really acetic acid? Name Per What part of the solution is really HC2H3O2 aka CH3COOH? What is the Ka of acetic acid? Introduction: The acid in vinegar is acetic
Polyethylene Glycol (PEG), High Sensitive ELISA
 K-ASSAY Polyethylene Glycol (PEG), High Sensitive ELISA For the high sensitive quantitative determination of PEG and PEGylated proteins in serum or plasma Cat. No. KT-657 For Research Use Only. 1 K-ASSAY
K-ASSAY Polyethylene Glycol (PEG), High Sensitive ELISA For the high sensitive quantitative determination of PEG and PEGylated proteins in serum or plasma Cat. No. KT-657 For Research Use Only. 1 K-ASSAY
Supernatant: The liquid layer lying above the solid layer after a precipitation reaction occurs.
 Limiting Reagent Introduction The quantities of substances involved in a chemical reaction represented by a balanced equation are often referred to as stoichiometric amounts. Solution stoichiometry is
Limiting Reagent Introduction The quantities of substances involved in a chemical reaction represented by a balanced equation are often referred to as stoichiometric amounts. Solution stoichiometry is
Experiment 7: ACID-BASE TITRATION: STANDARDIZATION OF A SOLUTION
 Experiment 7: ACID-BASE TITRATION: STANDARDIZATION OF A SOLUTION Purpose: Determine molarity of a solution of unknown concentration by performing acid-base titrations Performance Goals: Apply the concepts
Experiment 7: ACID-BASE TITRATION: STANDARDIZATION OF A SOLUTION Purpose: Determine molarity of a solution of unknown concentration by performing acid-base titrations Performance Goals: Apply the concepts
Materials Per Class Per Bench. 50 ml beakers 6 1. Hole punch 6 1. Forceps 6 1. Timers or a clock with second hand 6 1
 Photosynthesis Materials Per Class Per Bench 1% solution of sodium bicarbonate (NaHCO 3 ) (by adding approximately 1g sodium bicarbonate to 100 ml DI water). Light sources, 60 watt bulb or higher 3 or
Photosynthesis Materials Per Class Per Bench 1% solution of sodium bicarbonate (NaHCO 3 ) (by adding approximately 1g sodium bicarbonate to 100 ml DI water). Light sources, 60 watt bulb or higher 3 or
Electrolysis SCIENCE TOPICS PROCESS SKILLS VOCABULARY
 SIDE DISPLAY Electrolysis Visitors observe two glass tubes containing water. Electrodes are passing an electric current through the water. Oxygen gas is formed in the tube connected to the positive electrode.
SIDE DISPLAY Electrolysis Visitors observe two glass tubes containing water. Electrodes are passing an electric current through the water. Oxygen gas is formed in the tube connected to the positive electrode.
Non-Interfering Protein Assay Kit Cat. No
 Visit our interactive pathways at /pathways User Protocol 488250 Rev. 5 January 2006 RFH Page 1 of 1 Non-Interfering Protein Assay Kit Cat. No. 488250 Note that this user protocol is not lot-specific and
Visit our interactive pathways at /pathways User Protocol 488250 Rev. 5 January 2006 RFH Page 1 of 1 Non-Interfering Protein Assay Kit Cat. No. 488250 Note that this user protocol is not lot-specific and
O H 3 O 1 1 A. O 1 1 OH (K w
 CHAPTER 8 Acid Base Titration Curves Objectives The objectives of this experiment are to: Understand the titration curves for the following solutions: a strong acid: hydrochloric acid, HCl. a weak acid:
CHAPTER 8 Acid Base Titration Curves Objectives The objectives of this experiment are to: Understand the titration curves for the following solutions: a strong acid: hydrochloric acid, HCl. a weak acid:
LAB. FACTORS INFLUENCING ENZYME ACTIVITY
 AP Biology Date LAB. FACTORS INFLUENCING ENZYME ACTIVITY Background Enzymes are biological catalysts capable of speeding up chemical reactions by lowering activation energy. One benefit of enzyme catalysts
AP Biology Date LAB. FACTORS INFLUENCING ENZYME ACTIVITY Background Enzymes are biological catalysts capable of speeding up chemical reactions by lowering activation energy. One benefit of enzyme catalysts
Measurements with Ion Selective Electrodes: Determination of Fluoride in Toothpaste
 Experiment ISE: Measurements with Ion Selective Electrodes: Determination of Fluoride in Toothpaste 67 You have been hired by the government to check the fluoride concentration labelling on some major
Experiment ISE: Measurements with Ion Selective Electrodes: Determination of Fluoride in Toothpaste 67 You have been hired by the government to check the fluoride concentration labelling on some major
By contrast, solubility equilibrium reactions are written from the perspective of the solid reactant dissolving into ions
 LAD F.2 (pg 1 of 8) Ksp Solubility Product for Calcium Hydroxide Name Per Introduction Most solubility equilibrium investigated in this course involve ionic compounds as opposed to molecular compounds.
LAD F.2 (pg 1 of 8) Ksp Solubility Product for Calcium Hydroxide Name Per Introduction Most solubility equilibrium investigated in this course involve ionic compounds as opposed to molecular compounds.
Photosynthesis: How do plants get engery? Teacher Version
 Photosynthesis: How do plants get engery? Teacher Version In this lab, students explore the process of photosynthesis in spinach leaves. As oxygen is produced, the density of the leaves change and they
Photosynthesis: How do plants get engery? Teacher Version In this lab, students explore the process of photosynthesis in spinach leaves. As oxygen is produced, the density of the leaves change and they
2/22/2019 NEW UNIT! Chemical Interactions. Atomic Basics #19
 NEW UNIT! Chemical Interactions Atomic Basics #19 1 Vocabulary: Matter: Anything that has mass and takes up space. Atom: the smallest particle of matter. Element: A pure substance made up of only one type
NEW UNIT! Chemical Interactions Atomic Basics #19 1 Vocabulary: Matter: Anything that has mass and takes up space. Atom: the smallest particle of matter. Element: A pure substance made up of only one type
Kinetics: Factors that Affect Rates of Chemical Reactions
 Objective- Study several factors that influence chemical reaction rates, including: 1. Concentration 2. The nature of the chemical reactants 3. Area in a heterogeneous reaction 4. The temperature of the
Objective- Study several factors that influence chemical reaction rates, including: 1. Concentration 2. The nature of the chemical reactants 3. Area in a heterogeneous reaction 4. The temperature of the
ACID-BASE TITRATION (MICROSCALE)
 ACID-BASE TITRATION (MICROSCALE) LAB PH 4.PALM From Science with Handhelds, Vernier Software & Technology, 2002. INTRODUCTION Acids and bases represent a major class of chemical substances. We encounter
ACID-BASE TITRATION (MICROSCALE) LAB PH 4.PALM From Science with Handhelds, Vernier Software & Technology, 2002. INTRODUCTION Acids and bases represent a major class of chemical substances. We encounter
Conductometric Titration & Gravimetric Determination of a Precipitate
 Conductometric Titration & Gravimetric Determination of a Precipitate Experiment 9 In this experiment, you will monitor conductivity during the reaction between sulfuric acid, H2SO4, and barium hydroxide,
Conductometric Titration & Gravimetric Determination of a Precipitate Experiment 9 In this experiment, you will monitor conductivity during the reaction between sulfuric acid, H2SO4, and barium hydroxide,
Working in the Chemistry Laboratory
 Working in the Chemistry Laboratory Accelerated Chemistry I Introduction: One of the most important components of your chemistry course is the laboratory experience. Perhaps you have done experiments in
Working in the Chemistry Laboratory Accelerated Chemistry I Introduction: One of the most important components of your chemistry course is the laboratory experience. Perhaps you have done experiments in
University of Maryland Department of Physics
 University of Maryland Department of Physics Physics 131 Fall 2011 Sample problems Exam 2 (25 points) Using a rope of negligible mass, a box is pulled along a horizontal surface as shown in the figure
University of Maryland Department of Physics Physics 131 Fall 2011 Sample problems Exam 2 (25 points) Using a rope of negligible mass, a box is pulled along a horizontal surface as shown in the figure
Bovine FSH(Follicle Stimulating Hormone) ELISA Kit
 Bovine FSH(Follicle Stimulating Hormone) ELISA Kit Catalogue No.: EB0141 Size: 48T/96T Reactivity: Bovine Detection Range: 1.563-100mIU/ml Sensitivity:
Bovine FSH(Follicle Stimulating Hormone) ELISA Kit Catalogue No.: EB0141 Size: 48T/96T Reactivity: Bovine Detection Range: 1.563-100mIU/ml Sensitivity:
CHAPTER 4 THERMAL CONDUCTIVITY AND VISCOSITY MEASUREMENTS
 50 CHAPTER 4 THERMAL CONDUCTIVITY AND VISCOSITY MEASUREMENTS 4.1 INTRODUCTION In the development of any energy-efficient heat transfer fluids for enhanced heat transfer performance, in practical applications,
50 CHAPTER 4 THERMAL CONDUCTIVITY AND VISCOSITY MEASUREMENTS 4.1 INTRODUCTION In the development of any energy-efficient heat transfer fluids for enhanced heat transfer performance, in practical applications,
Atoms. - Proton - Neutron. - Electron
 Chemistry of Life Atoms - The basic unit of matter is called an Atom - Atoms are incredibly small, but despite its extremely small size, an atom contains subatomic particles that are even smaller - Three
Chemistry of Life Atoms - The basic unit of matter is called an Atom - Atoms are incredibly small, but despite its extremely small size, an atom contains subatomic particles that are even smaller - Three
Lab 3: Transpiration. 1 Purpose. BIO124 Plant Science Lab 3 Transpiration 1
 1 Purpose The goals of this lab are to (1) observe water movement against gravity from stems to leaves of plants and (2) investigate environmental factors that regulate the rate of transpiration. Introduction
1 Purpose The goals of this lab are to (1) observe water movement against gravity from stems to leaves of plants and (2) investigate environmental factors that regulate the rate of transpiration. Introduction
Transpiration Lab. Introduction
 Transpiration Lab Name Introduction The amount of water needed daily by plants for the growth and maintenance of tissues is small in comparison to the amount that is lost through the process of transpiration
Transpiration Lab Name Introduction The amount of water needed daily by plants for the growth and maintenance of tissues is small in comparison to the amount that is lost through the process of transpiration
Modified Adams Assay for Phenolics in Wine
 Modified Adams Assay for Phenolics in Wine 1. Total Iron-Reactive Phenolics THIS VALUE WILL DETERMINE DILUTIONS FOR TANNIN & POLYMERIC PIGMENT ANALYSES 1.1 Into a reduced volume cuvette, pipette in the
Modified Adams Assay for Phenolics in Wine 1. Total Iron-Reactive Phenolics THIS VALUE WILL DETERMINE DILUTIONS FOR TANNIN & POLYMERIC PIGMENT ANALYSES 1.1 Into a reduced volume cuvette, pipette in the
Hepatitis B virus IgM ELISA Kit
 Hepatitis B virus IgM ELISA Kit Catalog Number KA0289 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay...
Hepatitis B virus IgM ELISA Kit Catalog Number KA0289 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay...
TF (Bovine) ELISA Kit
 TF (Bovine) ELISA Kit Catalog Number KA2047 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay... 3 General
TF (Bovine) ELISA Kit Catalog Number KA2047 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay... 3 General
