Welcome to Week 5. Chapter 9 - Binding, Structure, and Diversity. 9.1 Intermolecular Forces. Starting week five video. Introduction to Chapter 9
|
|
|
- Dominick Martin
- 5 years ago
- Views:
Transcription
1 Welcome to Week 5 Starting week five video Please watch the online video (49 seconds). Chapter 9 - Binding, Structure, and Diversity Introduction to Chapter 9 Chapter 9 contains six subsections. Intermolecular Forces Case Study - Stromelysin Drug-Target Complementarity Molecular Diversity Molecular Libraries Building Libraries Upon completing this chapter, you should understand how drugs bind a target and how to determine the energy of binding. You should gain a preliminary idea of how to control the shape of a molecule in order to maximize available drug-target interactions. You should realize the challenge of discovering active molecules within the immense number of possible drug molecules and what tools are available to drug companies to explore drug space. 9.1 Intermolecular Forces Binding energy video Please watch the online video (10 minutes, 26 seconds). Clarifications and corrections At approximately the 4:11 mark, the video states that 17x10-3 M is equal to 1.7x10-4 M instead of 1.7x10-2 M.
2 A condensed summary of this video can be found in the Video summary page. Henderson-Hasselbalch equation Background: Hydrogen and ionic bonds are very important for drug-target binding. These bonds can be highly dependent upon ph. Instructions: Read the passage below concerning the Henderson-Hasselbalch equation and the pk a of common functional groups. Use the information to answer the questions that follow. Learning Goals: To review usage of the Henderson-Hasselbalch equation and understand how ph influences the availability of hydrogen and ionic bonds between a target and a drug. An acid (H-A) can react reversibly with water to form a conjugate base (A - ) and hydronium ion (H 3O + ). The equilibrium constant (K) for this reaction can be expressed in the standard way - the product of the concentrations of the products divided by the product of the concentrations of the reagents. Because the concentration of water is virtually constant at around 55 M, the term [H 2O] is combined with K to define a new constant, K a. While K a is a constant, the ratio of the acid and conjugate base in solution can be affected by the ph of the medium. A lower ph, with a raised [H 3O + ], favors H-A over A -. A higher ph, with a lowered [H 3O + ], favors more A -. The Henderson-Hasselbalch equation quantitatively relates how ph affects the equilibrium ratio of A - and H-A for an acid with a known K a. Remember that pk a = log K a. Henderson-Hasselbalch equation
3 To use the Henderson-Hasselbalch equation, one needs to know the pk a of different functional groups. pk a values for a handful of common functional groups encountered in drugs are shown in the table below. A much more comprehensive list can be found here ( acid (name) conjugate base (name) pk a H 3O + (hydronium) H 2O (water) 1.7 PhCO 2H (benzoic acid) PhCO 2 (benzoate) PhNH 3 (anilinium) PhNH 2 (aniline) 4.6 CH 3CO 2H (acetic acid) CH 3CO 2 (acetate) 4.8 (pyridinium) (pyridine) 5.1 (imidazolium) (imidazole) NH 4 (ammonium) NH 3 (ammonia) 9.2 PhOH (phenol) PhO (phenolate) 10.0 H 2O (water) HO (hydroxide) 15.7 For reference, the ph of some different regions in the body are listed below. blood stomach - as low as 1 small intestine extracellular fluid intracellular fluid Please complete the online exercise.
4 Calculating binding energies Background: The binding energy of a drug-target complex can be calculated with the equation below. If kcal/mol*k is used for R, then the energy is calculated with units of kcal/mol. Temperature (T) is usually 298 K. Instructions: Use the equation above to answer the questions that follow. Learning Goal: To practice calculating binding energies between a drug and target. Please complete the online exercise. FAQ, help, and tips Binding energy video Why does the course switch from natural logarithms (ln or log e) to base-10 logarithms (log 10)? In treatments of pharmacokinetics, the relationship of C p and time is normally presented in a natural logarithm form (lnc p vs. time). In contrast, in discussions of binding energies, the use of base-10 logarithms is more standard. Why are binding energies reported in kcal/mol instead of kj/mol? Medicinal chemistry is dominated by organic chemists, who tend to favor energies in terms of kcal/mol instead of kj/mol. Is there a reference for the 0.03 kcal/mol/å 2? Yes. The original reference for that value is... Chothia, C. Hydrophobic bonding and accessible surface area in proteins. Nature 1974, 248,
5 Calculating binding energies What temperature should be used for determining binding energies? Binding energies are normally determined through in vitro tests, which are performed at room temperature. Room temperature is close to 298 K. Here is a link to an article ( that goes into much greater detail on assays that measure binding energies. 9.2 Case Study - Stromelysin Stromelysin video Please watch the online video (8 minutes, 30 seconds). A condensed summary of this video can be found in the Video summary page. More stromelysin inhibitors Background: The binding energy of a drug-target complex can be calculated with the equation below. If kcal/mol*k is used for R, then the energy is calculated with units of kcal/mol. Temperature (T) is usually 298 K. Instructions: Use the equation above to answer the questions that follow concerning stromelysin inhibitors. Learning Goal: To practice calculating binding energies between an enzyme and its inhibitor. Please complete the online exercise.
6 FAQ, help, and tips Stromelysin video How can binding be measured on compounds with such high K D values? Weak binding molecules require specialized techniques for determining their K D values. Examples include surface plasmon resonance and saturation transfer difference NMR spectroscopy. More stromelysin inhibitors Please clarify the relationship between the K values, binding energies, and IC 50. First, there are two possible K values. Both are related to the complexation equilibrium of a drug and its target (an enzyme or receptor). The equilibrium can be defined in two directions. If the drug and target are considered to be the starting materials, then the corresponding equilibrium constant is K A - the association equilibrium constant. If the drug-target complex is the starting material, then the equilibrium constant is K D - the dissociation equilibrium constant. In drug discovery, the equilibrium constant of interest is almost always K D. A low value for K D indicates that the equilibrium favors the drug-target complex, and therefore binding between the drug and target is favorable (large, negative value for ΔG). K i values also dissociation equilibrium constants. K i values specifically refer to molecules that inhibit or block an enzyme or receptor. Just as with K D values, smaller K i values indicate an inhibitor that binds strongly to the target (large, negative value for ΔG). For a full agonist that binds a receptor, the K D of the ligand-receptor complex corresponds to the EC 50 of the ligand (the concentration of ligand required to affect a 50% maximal response from the receptor). Because K D = EC 50, it can be tempting to think that K i = IC 50. The equations look the same, so perhaps they must both be true. The relationship, K i = IC 50, however is not true. IC 50 is the concentration of an inhibitor that is required to reduce a receptor's response or an enzyme's rate of reaction by 50%. IC 50, however, varies based on how much of the receptor's agonist or enzyme's substrate is present. If more agonist or substrate is present, then more
7 inhibitor will be required (IC 50 will be higher). In contrast, K i is a constant property of the inhibitortarget complex. Remember that the Cheng-Prussoff equation allows the determination of K i from an IC 50 value as long as one knows two things. First is the concentration of the agonist ([L]) used to determine the IC 50 value. Second is the K D of the agonist for the receptor. If the target is an enzyme, then one needs to know the concentration of the substrate ([S]) and the K m of the substrate for the enzyme. Can you provide a reference to the stromelysin research? Here is a link to a copy of one of the early articles from the Abbott researchers. ( 9.3 Drug-Target Complementarity Pharmacophores revisited video Please watch the online video (7 minutes, 32 seconds). A condensed summary of this video can be found in the Video summary page. Rotatable bonds Background: Rotatable bonds increase the conformational flexibility of a molecule and minimize the probability that the functional groups in a molecule match the desired pharmacophore. Instructions: Read the passage below on identifying rotatable bonds. Use the information to answer the questions that follow. Learning Goal: To learn how to identify which bonds in a molecule qualify as rotatable. No universally accepted rules exist on defining rotatable bonds. Each set of definitions has its loop holes and problems. Regardless, most methods include the rules shown below.
8 Bonds that are not rotatable... non single bonds bonds to hydrogen and other monovalent atoms (halogens) ring bonds bonds to terminal atoms, including CH 3, NH 2, and OH the C-N bond between a carbonyl and amide nitrogen (also goes for the C-N bond in thioamides and the S-N bond in sulfonamides) bonds connecting two aromatic rings with collectively three or more ortho substituents bonds connected to terminal triple bonds, including bonds to cyano groups Under these rules, naproxen has only three rotatable bonds (bolded below). Duloxetine has six (bolded below). Please complete the online exercise. FAQ, help, and tips Pharmacophores revisited video How can researchers possibly know what groups are required in the pharmacophore? Determining the pharmacophore of a drug may seem like an insurmountable organic chemistry puzzle, but researchers often have considerable information at their disposal. The biology group will have intensely studied any target, whether an enzyme or receptor. For enzymes, the substrate will almost certainly be known. The same can be said for the endogenous ligand of a receptor. If the substrate or natural ligand is known, then the medicinal chemistry group has a handle on the type of structure that has a high affinity for a binding pocket on the target. That information might be enough for the discovery team to search intelligently for promising compounds.
9 Drug discovery teams often also have x-ray structural information on a target. An x-ray structure allows the team to visualize in three-dimensions the contours of different binding pockets of the target. If an x-ray structure of the target is available, often an x-ray structure of the target complexed with a second molecule will also be at hand. An x-ray of a target that has co-crystallized with another molecule allows the discovery group to see precisely how the functional groups of the bound molecule interact with the target. Potential hydrophobic interactions and hydrogen bonding interactions can be clear. Knowing these interactions can allow the discovery group to hypothesize the structure of the pharmacophore and begin to plan how binding might be improved in subsequent candidate molecules. 9.4 Molecular Diversity Numbers game video Please watch the online video (7 minutes, 55 seconds). A condensed summary of this video can be found in the Video summary page. Privileged structures Background: Molecular space for potential drug molecules is indescribably diverse. Instructions: Read the passage below about certain types of structures and substructures that repeatedly appear in many different types of drug. Learning Goal: To understand the concept of privileged structures. While drug space is incredibly diverse, a handful of common structures and molecular fragments can be found regularly among active sets of molecules. These compounds and fragments have become known as privileged structures because of their seemingly universal ability to bind protein targets.
10 One such privileged structure is the diphenylmethane subunit. The subunit can be seen in numerous compounds that bind a variety of different targets. Privileged structures are both good and bad for drug discovery. On the good side, privileged structures help researchers focus on molecular scaffolds that are more likely to show activity. The drug discovery team can focus on promising compounds and hopefully avoid the less interesting structures. On the bad side, single compounds that contain privileged structures may show activity against multiple targets. Compounds that bind multiple targets often cause side effects. Such compounds are sometimes labeled as promiscuous. Another issue with privileged structures is that they have received considerable research attention. Patenting a compound with a privileged structure can be a challenge because so many similar structures have already been patented. Promiscuity revealed Background: Privileged structures are molecular scaffolds that tend to show activity against a broad range of biological targets. Such compounds are sometimes said to have promiscuous activity. Instructions: Read about Molinspiration's Predict Bioactivity function for JSME and use this function to rank the promiscuity of a set of privileged scaffolds. Learning Goal: To gain exposure to emerging biological activity prediction tools. Molinspiration, a site that we have used for determining whether a compound satisfies Lipinski's rules, has coupled a Predict Bioactivity function with the JSME tool. With this functionality, a user may predict the activity of a structure against six different types of commonly encountered drug targets, including GPCRs, ion channels, kinases, nuclear receptors, proteases.
11 Molinspiration's page is shown below with a structure of a known protease inhibitor, captopril, drawn in the JSME tool. Captopril is used to manage high blood pressure. Once the "Predict Bioactivity" button is clicked, the Molinspiration engine checks the properties of the structure against a library of molecules with known biological activity and predicts the activity of the submitted structure. The predicted activities are then shown to the user. Activities are predicted through a numerical score, larger and positive values indicate higher predicted activity. If a molecule is predicted to have activity against a target, then the target is shaded light green (moderate activity) or dark green (strong activity). In the case of captopril, the Molinspiration tool successfully identifies the known protease activity of structure. With this webpage, one can make educated predictions on what type of targets might be bound by molecule. Please complete the online exercise.
12 FAQ, help, and tips Numbers game video What is the original Guida reference? Here is a link to the original article. ( The full text is unfortunately not freely available. Behind the numbers Can edx accept answers in scientific notation in other formats? The question suggests the following format. 6.8*10^2 Another valid and simpler format is shown below. 6.8e2 Privileged structures Are there reviews on privileged structures? One review on privileged structures can be found through this link. ( CurrOpinChemBiol_2010.pdf) 9.5 Molecular Libraries Molecule collections video Please watch the online video (6 minutes 56 seconds). A condensed summary of this video can be found in the Video summary page.
13 High-throughput screening Background: Potential drug space is immense with an estimated number of molecules of Compound libraries, although nowhere near the same size as drug space, are regardless very large and may include 1,000,000 or more compounds. Instructions: Read the text below and the accompanying Wikipedia entry concerning the rapid testing of molecular libraries. Learning Goal: To understand how the large number of molecules in compound libraries can be quickly and inexpensively tested for biological activity. For molecular libraries to be useful to a drug company, there must exist a method for quickly and inexpensively testing the activity of each compound in the library. Fortunately there is. That method is called high-throughput screening or HTS. HTS involves the automated testing of molecules in a quick, inexpensive, in vitro assay. The process relies upon robotic equipment to perform the screen reproducibly. With such a method, a pharmaceutical company can screen an entire large library, which may include a million or more compounds, in around a week in a particular screen. Therefore, in a fairly short period of time, hits for a specific target can be identified. The entry for high-throughput screening in Wikipedia provides some more details on precisely how the process is automated. The statistical and emerging technology discussions in the article are beyond the scope of this course, but they do reveal interesting facets of HTS. HTS and academia Background: High-throughput screening (HTS) is an automated, quick method for gaining preliminary activity information on molecules in a compound library. HTS has traditionally been a technique only available to pharmaceutical companies. Instructions: Read the article below. Use the information in the article to answer the questions that follow. Learning Goal: To understand how the drug discovery process is becoming increasingly available to groups outside the traditional pharmaceutical industry. A recent report in the journal Nature Methods notes the growing movement of academic laboratories screening their own molecular libraries. ( Please complete the online exercise.
14 FAQ, help, and tips HTS and academia Exactly how does HTS make drug discovery faster? In drug discovery, sometimes the research team has little or no information to help find a molecule that binds the desired target. The only option is to test individual molecules for binding a target in an assay. Traditional biochemical assays, when performed individually, can be very time intensive. When performed in an automated fashion with robotic assistance, the time required for the assays can be greatly reduced. In this manner, the automated assays, called high-throughput screens, can accelerate the discovery of molecules with promising target binding 9.6 Building Libraries Combinatorial chemistry video Please watch the online video (7 minutes 10 seconds). A condensed summary of this video can be found in the Video summary page. Ugi reaction Background: Reactions that can quickly and easily assemble large numbers of diverse molecules are highly sought after in combinatorial chemistry. Instructions: Read passage below on the Ugi reaction and answer the question that follows. Learning Goal: To learn about a reaction that is commonly exploited in combinatorial chemistry. The Ugi reaction involves the reaction of four different starting materials in a single reaction vessel. The starting materials are a carboxylic acid (1), primary amine (2), aldehyde (3), and isocyanide (4).
15 The simplicity of the Ugi reaction and the widespread availability of the starting materials make the reaction very popular starting point in preparing molecular scaffolds in combinatorial chemistry. Please complete the online exercise. Combichem challenges Background: In the early 1990s combinatorial chemistry was hoped to accelerate greatly the rate at which new drugs could be discovered. The benefits of combinatorial chemistry did not however materialize as expected. Instructions: Read the Chemistry and Engineering News article linked below. Use the information to answer the questions that follow. Learning Goal: To learn about a reaction that is commonly exploited in combinatorial chemistry. Although over 15 years old, an article from Chemistry and Engineering News, a trade magazine published by the American Chemical Society, captures the early sentiments of a booming field called combinatorial chemistry. Even in 1998 in the face of great enthusiasm, the promises of combinatorial chemistry were already being questioned. The article nicely captures both sides of the story. ( Please complete the online exercise. FAQ, help, and tips Combinatorial chemistry video Please provide an additional reference on high-throughput screening. The Wikipedia article on HTS is very dense. A freely available article ( provides a good, although long summary of the field. The language of the article fits well with the terminology that has been covered in this course.
16 Combi chem challenges Is there any other literature that more fully describes limitations of combinatorial chemistry libraries for drug discovery? Here is an article by Lipinski that goes into some detail about combi chem library problems. (
Receptor Based Drug Design (1)
 Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
COMBINATORIAL CHEMISTRY: CURRENT APPROACH
 COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
Early Stages of Drug Discovery in the Pharmaceutical Industry
 Early Stages of Drug Discovery in the Pharmaceutical Industry Daniel Seeliger / Jan Kriegl, Discovery Research, Boehringer Ingelheim September 29, 2016 Historical Drug Discovery From Accidential Discovery
Early Stages of Drug Discovery in the Pharmaceutical Industry Daniel Seeliger / Jan Kriegl, Discovery Research, Boehringer Ingelheim September 29, 2016 Historical Drug Discovery From Accidential Discovery
Introduction to Chemoinformatics and Drug Discovery
 Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
EMPIRICAL VS. RATIONAL METHODS OF DISCOVERING NEW DRUGS
 EMPIRICAL VS. RATIONAL METHODS OF DISCOVERING NEW DRUGS PETER GUND Pharmacopeia Inc., CN 5350 Princeton, NJ 08543, USA pgund@pharmacop.com Empirical and theoretical approaches to drug discovery have often
EMPIRICAL VS. RATIONAL METHODS OF DISCOVERING NEW DRUGS PETER GUND Pharmacopeia Inc., CN 5350 Princeton, NJ 08543, USA pgund@pharmacop.com Empirical and theoretical approaches to drug discovery have often
Computational chemical biology to address non-traditional drug targets. John Karanicolas
 Computational chemical biology to address non-traditional drug targets John Karanicolas Our computational toolbox Structure-based approaches Ligand-based approaches Detailed MD simulations 2D fingerprints
Computational chemical biology to address non-traditional drug targets John Karanicolas Our computational toolbox Structure-based approaches Ligand-based approaches Detailed MD simulations 2D fingerprints
Structural biology and drug design: An overview
 Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
CHEM 4170 Problem Set #1
 CHEM 4170 Problem Set #1 0. Work problems 1-7 at the end of Chapter ne and problems 1, 3, 4, 5, 8, 10, 12, 17, 18, 19, 22, 24, and 25 at the end of Chapter Two and problem 1 at the end of Chapter Three
CHEM 4170 Problem Set #1 0. Work problems 1-7 at the end of Chapter ne and problems 1, 3, 4, 5, 8, 10, 12, 17, 18, 19, 22, 24, and 25 at the end of Chapter Two and problem 1 at the end of Chapter Three
Properties of Amines
 Properties of Amines 1. Boiling Point and Water Solubility It is instructive to compare the boiling points and water solubility of amines with those of corresponding alcohols and ethers. The dominant factor
Properties of Amines 1. Boiling Point and Water Solubility It is instructive to compare the boiling points and water solubility of amines with those of corresponding alcohols and ethers. The dominant factor
CSD. Unlock value from crystal structure information in the CSD
 CSD CSD-System Unlock value from crystal structure information in the CSD The Cambridge Structural Database (CSD) is the world s most comprehensive and up-todate knowledge base of crystal structure data,
CSD CSD-System Unlock value from crystal structure information in the CSD The Cambridge Structural Database (CSD) is the world s most comprehensive and up-todate knowledge base of crystal structure data,
Statistical concepts in QSAR.
 Statistical concepts in QSAR. Computational chemistry represents molecular structures as a numerical models and simulates their behavior with the equations of quantum and classical physics. Available programs
Statistical concepts in QSAR. Computational chemistry represents molecular structures as a numerical models and simulates their behavior with the equations of quantum and classical physics. Available programs
Biologically Relevant Molecular Comparisons. Mark Mackey
 Biologically Relevant Molecular Comparisons Mark Mackey Agenda > Cresset Technology > Cresset Products > FieldStere > FieldScreen > FieldAlign > FieldTemplater > Cresset and Knime About Cresset > Specialist
Biologically Relevant Molecular Comparisons Mark Mackey Agenda > Cresset Technology > Cresset Products > FieldStere > FieldScreen > FieldAlign > FieldTemplater > Cresset and Knime About Cresset > Specialist
Ping-Chiang Lyu. Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University.
 Pharmacophore-based Drug design Ping-Chiang Lyu Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University 96/08/07 Outline Part I: Analysis The analytical
Pharmacophore-based Drug design Ping-Chiang Lyu Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University 96/08/07 Outline Part I: Analysis The analytical
In Silico Investigation of Off-Target Effects
 PHARMA & LIFE SCIENCES WHITEPAPER In Silico Investigation of Off-Target Effects STREAMLINING IN SILICO PROFILING In silico techniques require exhaustive data and sophisticated, well-structured informatics
PHARMA & LIFE SCIENCES WHITEPAPER In Silico Investigation of Off-Target Effects STREAMLINING IN SILICO PROFILING In silico techniques require exhaustive data and sophisticated, well-structured informatics
1. (18) Multiple choice questions. Please place your answer on the line preceding each question.
 CEM 5720 ame KEY Exam 2 ctober 21, 2015 Read all questions carefully and attempt those questions you are sure of first. Remember to proof your work; art work carries as much importance as written responses.
CEM 5720 ame KEY Exam 2 ctober 21, 2015 Read all questions carefully and attempt those questions you are sure of first. Remember to proof your work; art work carries as much importance as written responses.
In silico pharmacology for drug discovery
 In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
COMBINATORIAL CHEMISTRY IN A HISTORICAL PERSPECTIVE
 NUE FEATURE T R A N S F O R M I N G C H A L L E N G E S I N T O M E D I C I N E Nuevolution Feature no. 1 October 2015 Technical Information COMBINATORIAL CHEMISTRY IN A HISTORICAL PERSPECTIVE A PROMISING
NUE FEATURE T R A N S F O R M I N G C H A L L E N G E S I N T O M E D I C I N E Nuevolution Feature no. 1 October 2015 Technical Information COMBINATORIAL CHEMISTRY IN A HISTORICAL PERSPECTIVE A PROMISING
Using AutoDock for Virtual Screening
 Using AutoDock for Virtual Screening CUHK Croucher ASI Workshop 2011 Stefano Forli, PhD Prof. Arthur J. Olson, Ph.D Molecular Graphics Lab Screening and Virtual Screening The ultimate tool for identifying
Using AutoDock for Virtual Screening CUHK Croucher ASI Workshop 2011 Stefano Forli, PhD Prof. Arthur J. Olson, Ph.D Molecular Graphics Lab Screening and Virtual Screening The ultimate tool for identifying
A. Reaction Mechanisms and Catalysis (1) proximity effect (2) acid-base catalysts (3) electrostatic (4) functional groups (5) structural flexibility
 (P&S Ch 5; Fer Ch 2, 9; Palm Ch 10,11; Zub Ch 9) A. Reaction Mechanisms and Catalysis (1) proximity effect (2) acid-base catalysts (3) electrostatic (4) functional groups (5) structural flexibility B.
(P&S Ch 5; Fer Ch 2, 9; Palm Ch 10,11; Zub Ch 9) A. Reaction Mechanisms and Catalysis (1) proximity effect (2) acid-base catalysts (3) electrostatic (4) functional groups (5) structural flexibility B.
October 6 University Faculty of pharmacy Computer Aided Drug Design Unit
 October 6 University Faculty of pharmacy Computer Aided Drug Design Unit CADD@O6U.edu.eg CADD Computer-Aided Drug Design Unit The development of new drugs is no longer a process of trial and error or strokes
October 6 University Faculty of pharmacy Computer Aided Drug Design Unit CADD@O6U.edu.eg CADD Computer-Aided Drug Design Unit The development of new drugs is no longer a process of trial and error or strokes
Retrieving hits through in silico screening and expert assessment M. N. Drwal a,b and R. Griffith a
 Retrieving hits through in silico screening and expert assessment M.. Drwal a,b and R. Griffith a a: School of Medical Sciences/Pharmacology, USW, Sydney, Australia b: Charité Berlin, Germany Abstract:
Retrieving hits through in silico screening and expert assessment M.. Drwal a,b and R. Griffith a a: School of Medical Sciences/Pharmacology, USW, Sydney, Australia b: Charité Berlin, Germany Abstract:
Introduction. OntoChem
 Introduction ntochem Providing drug discovery knowledge & small molecules... Supporting the task of medicinal chemistry Allows selecting best possible small molecule starting point From target to leads
Introduction ntochem Providing drug discovery knowledge & small molecules... Supporting the task of medicinal chemistry Allows selecting best possible small molecule starting point From target to leads
CHEM 109A Organic Chemistry
 CHEM 109A Organic Chemistry https://labs.chem.ucsb.edu/zakarian/armen/courses.html Chapter 2 Acids and Bases Central to Understanding Organic Chemistry Draw the conjugate acid of each of the following:
CHEM 109A Organic Chemistry https://labs.chem.ucsb.edu/zakarian/armen/courses.html Chapter 2 Acids and Bases Central to Understanding Organic Chemistry Draw the conjugate acid of each of the following:
Chapter 8. Acidity, Basicity and pk a
 Chapter 8 Acidity, Basicity and pk a p182 In this reaction water is acting as a base, according to our definition above, by accepting a proton from HCl which in turn is acting as an acid by donating a
Chapter 8 Acidity, Basicity and pk a p182 In this reaction water is acting as a base, according to our definition above, by accepting a proton from HCl which in turn is acting as an acid by donating a
Chapter 20: Carboxylic Acids and Nitriles شیمی آلی 2
 Chapter 20: Carboxylic Acids and Nitriles شیمی آلی 2 Dr M. Mehrdad University of Guilan, Department of Chemistry, Rasht, Iran m-mehrdad@guilan.ac.ir Based on McMurry s Organic Chemistry, 7 th edition The
Chapter 20: Carboxylic Acids and Nitriles شیمی آلی 2 Dr M. Mehrdad University of Guilan, Department of Chemistry, Rasht, Iran m-mehrdad@guilan.ac.ir Based on McMurry s Organic Chemistry, 7 th edition The
Chapter 8 Lecture Notes: Acids, Bases, and ph
 Educational Goals Chapter 8 Lecture Notes: Acids, Bases, and ph 1. Given a chemical equation, write the law of mass action. 2. Given the equilibrium constant (K eq ) for a reaction, predict whether the
Educational Goals Chapter 8 Lecture Notes: Acids, Bases, and ph 1. Given a chemical equation, write the law of mass action. 2. Given the equilibrium constant (K eq ) for a reaction, predict whether the
Identifying Interaction Hot Spots with SuperStar
 Identifying Interaction Hot Spots with SuperStar Version 1.0 November 2017 Table of Contents Identifying Interaction Hot Spots with SuperStar... 2 Case Study... 3 Introduction... 3 Generate SuperStar Maps
Identifying Interaction Hot Spots with SuperStar Version 1.0 November 2017 Table of Contents Identifying Interaction Hot Spots with SuperStar... 2 Case Study... 3 Introduction... 3 Generate SuperStar Maps
Full file at Chapter 2 Water: The Solvent for Biochemical Reactions
 Chapter 2 Water: The Solvent for Biochemical Reactions SUMMARY Section 2.1 Summary Water is a polar molecule, with a partial negative charge on the oxygen and partial positive charges on the hydrogens.
Chapter 2 Water: The Solvent for Biochemical Reactions SUMMARY Section 2.1 Summary Water is a polar molecule, with a partial negative charge on the oxygen and partial positive charges on the hydrogens.
Concept review: Binding equilibria
 Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
Structure-based maximal affinity model predicts small-molecule druggability
 Structure-based maximal affinity model predicts small-molecule druggability Alan Cheng alan.cheng@amgen.com IMA Workshop (Jan 17, 2008) Druggability prediction Introduction Affinity model Some results
Structure-based maximal affinity model predicts small-molecule druggability Alan Cheng alan.cheng@amgen.com IMA Workshop (Jan 17, 2008) Druggability prediction Introduction Affinity model Some results
Development of Pharmacophore Model for Indeno[1,2-b]indoles as Human Protein Kinase CK2 Inhibitors and Database Mining
![Development of Pharmacophore Model for Indeno[1,2-b]indoles as Human Protein Kinase CK2 Inhibitors and Database Mining Development of Pharmacophore Model for Indeno[1,2-b]indoles as Human Protein Kinase CK2 Inhibitors and Database Mining](/thumbs/91/107031261.jpg) Development of Pharmacophore Model for Indeno[1,2-b]indoles as Human Protein Kinase CK2 Inhibitors and Database Mining Samer Haidar 1, Zouhair Bouaziz 2, Christelle Marminon 2, Tiomo Laitinen 3, Anti Poso
Development of Pharmacophore Model for Indeno[1,2-b]indoles as Human Protein Kinase CK2 Inhibitors and Database Mining Samer Haidar 1, Zouhair Bouaziz 2, Christelle Marminon 2, Tiomo Laitinen 3, Anti Poso
Design and Synthesis of the Comprehensive Fragment Library
 YOUR INNOVATIVE CHEMISTRY PARTNER IN DRUG DISCOVERY Design and Synthesis of the Comprehensive Fragment Library A 3D Enabled Library for Medicinal Chemistry Discovery Warren S Wade 1, Kuei-Lin Chang 1,
YOUR INNOVATIVE CHEMISTRY PARTNER IN DRUG DISCOVERY Design and Synthesis of the Comprehensive Fragment Library A 3D Enabled Library for Medicinal Chemistry Discovery Warren S Wade 1, Kuei-Lin Chang 1,
Dr. Sander B. Nabuurs. Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre
 Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
CSD. CSD-Enterprise. Access the CSD and ALL CCDC application software
 CSD CSD-Enterprise Access the CSD and ALL CCDC application software CSD-Enterprise brings it all: access to the Cambridge Structural Database (CSD), the world s comprehensive and up-to-date database of
CSD CSD-Enterprise Access the CSD and ALL CCDC application software CSD-Enterprise brings it all: access to the Cambridge Structural Database (CSD), the world s comprehensive and up-to-date database of
FRAGMENT SCREENING IN LEAD DISCOVERY BY WEAK AFFINITY CHROMATOGRAPHY (WAC )
 FRAGMENT SCREENING IN LEAD DISCOVERY BY WEAK AFFINITY CHROMATOGRAPHY (WAC ) SARomics Biostructures AB & Red Glead Discovery AB Medicon Village, Lund, Sweden Fragment-based lead discovery The basic idea:
FRAGMENT SCREENING IN LEAD DISCOVERY BY WEAK AFFINITY CHROMATOGRAPHY (WAC ) SARomics Biostructures AB & Red Glead Discovery AB Medicon Village, Lund, Sweden Fragment-based lead discovery The basic idea:
Data Quality Issues That Can Impact Drug Discovery
 Data Quality Issues That Can Impact Drug Discovery Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc, Sunnyvale, CA. 3 Royal Society of Chemistry,
Data Quality Issues That Can Impact Drug Discovery Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc, Sunnyvale, CA. 3 Royal Society of Chemistry,
Cheminformatics platform for drug discovery application
 EGI-InSPIRE Cheminformatics platform for drug discovery application Hsi-Kai, Wang Academic Sinica Grid Computing EGI User Forum, 13, April, 2011 1 Introduction to drug discovery Computing requirement of
EGI-InSPIRE Cheminformatics platform for drug discovery application Hsi-Kai, Wang Academic Sinica Grid Computing EGI User Forum, 13, April, 2011 1 Introduction to drug discovery Computing requirement of
Unit 1 ~ Learning Guide
 Unit 1 ~ Learning Guide Name: INSTRUCTIONS Complete the following notes and questions as you work through the related lessons. You are required to have this package completed BEFORE you write your unit
Unit 1 ~ Learning Guide Name: INSTRUCTIONS Complete the following notes and questions as you work through the related lessons. You are required to have this package completed BEFORE you write your unit
Carboxylic Acids and Nitriles
 Carboxylic Acids and Nitriles Why this Chapter? Carboxylic acids present in many industrial processes and most biological processes They are the starting materials from which other acyl derivatives are
Carboxylic Acids and Nitriles Why this Chapter? Carboxylic acids present in many industrial processes and most biological processes They are the starting materials from which other acyl derivatives are
Using Phase for Pharmacophore Modelling. 5th European Life Science Bootcamp March, 2017
 Using Phase for Pharmacophore Modelling 5th European Life Science Bootcamp March, 2017 Phase: Our Pharmacohore generation tool Significant improvements to Phase methods in 2016 New highly interactive interface
Using Phase for Pharmacophore Modelling 5th European Life Science Bootcamp March, 2017 Phase: Our Pharmacohore generation tool Significant improvements to Phase methods in 2016 New highly interactive interface
Classifying Organic Chemical Reactions
 Chemical Reactivity Organic chemistry encompasses a very large number of compounds ( many millions ), and our previous discussion and illustrations have focused on their structural characteristics. Now
Chemical Reactivity Organic chemistry encompasses a very large number of compounds ( many millions ), and our previous discussion and illustrations have focused on their structural characteristics. Now
AMRI COMPOUND LIBRARY CONSORTIUM: A NOVEL WAY TO FILL YOUR DRUG PIPELINE
 AMRI COMPOUD LIBRARY COSORTIUM: A OVEL WAY TO FILL YOUR DRUG PIPELIE Muralikrishna Valluri, PhD & Douglas B. Kitchen, PhD Summary The creation of high-quality, innovative small molecule leads is a continual
AMRI COMPOUD LIBRARY COSORTIUM: A OVEL WAY TO FILL YOUR DRUG PIPELIE Muralikrishna Valluri, PhD & Douglas B. Kitchen, PhD Summary The creation of high-quality, innovative small molecule leads is a continual
Dock Ligands from a 2D Molecule Sketch
 Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
More information can be found in Chapter 12 in your textbook for CHEM 3750/ 3770 and on pages in your laboratory manual.
 CHEM 3780 rganic Chemistry II Infrared Spectroscopy and Mass Spectrometry Review More information can be found in Chapter 12 in your textbook for CHEM 3750/ 3770 and on pages 13-28 in your laboratory manual.
CHEM 3780 rganic Chemistry II Infrared Spectroscopy and Mass Spectrometry Review More information can be found in Chapter 12 in your textbook for CHEM 3750/ 3770 and on pages 13-28 in your laboratory manual.
Ligand-receptor interactions
 University of Silesia, Katowice, Poland 11 22 March 2013 Ligand-receptor interactions Dr. Pavel Polishchuk A.V. Bogatsky Physico-Chemical Institute of National Academy of Sciences of Ukraine Odessa, Ukraine
University of Silesia, Katowice, Poland 11 22 March 2013 Ligand-receptor interactions Dr. Pavel Polishchuk A.V. Bogatsky Physico-Chemical Institute of National Academy of Sciences of Ukraine Odessa, Ukraine
EXPT. 9 DETERMINATION OF THE STRUCTURE OF AN ORGANIC COMPOUND USING UV, IR, NMR AND MASS SPECTRA
 EXPT. 9 DETERMINATION OF THE STRUCTURE OF AN ORGANIC COMPOUND USING UV, IR, NMR AND MASS SPECTRA Structure 9.1 Introduction Objectives 9.2 Principle 9.3 Requirements 9.4 Strategy for the Structure Elucidation
EXPT. 9 DETERMINATION OF THE STRUCTURE OF AN ORGANIC COMPOUND USING UV, IR, NMR AND MASS SPECTRA Structure 9.1 Introduction Objectives 9.2 Principle 9.3 Requirements 9.4 Strategy for the Structure Elucidation
Nonlinear QSAR and 3D QSAR
 onlinear QSAR and 3D QSAR Hugo Kubinyi Germany E-Mail kubinyi@t-online.de HomePage www.kubinyi.de onlinear Lipophilicity-Activity Relationships drug receptor Possible Reasons for onlinear Lipophilicity-Activity
onlinear QSAR and 3D QSAR Hugo Kubinyi Germany E-Mail kubinyi@t-online.de HomePage www.kubinyi.de onlinear Lipophilicity-Activity Relationships drug receptor Possible Reasons for onlinear Lipophilicity-Activity
Chemogenomic: Approaches to Rational Drug Design. Jonas Skjødt Møller
 Chemogenomic: Approaches to Rational Drug Design Jonas Skjødt Møller Chemogenomic Chemistry Biology Chemical biology Medical chemistry Chemical genetics Chemoinformatics Bioinformatics Chemoproteomics
Chemogenomic: Approaches to Rational Drug Design Jonas Skjødt Møller Chemogenomic Chemistry Biology Chemical biology Medical chemistry Chemical genetics Chemoinformatics Bioinformatics Chemoproteomics
Completions Multiple Enrollment in same semester. 2. Mode of Instruction (Hours per Unit are defaulted) Hegis Code(s) (Provided by the Dean)
 CALIFORNIA STATE UNIVERSITY CHANNEL ISLANDS COURSE MODIFICATION PROPOSAL Courses must be submitted by November 2, 2009, to make the next catalog (2010--2011) production DATE (CHANGE DATE EACH TIME REVISED):
CALIFORNIA STATE UNIVERSITY CHANNEL ISLANDS COURSE MODIFICATION PROPOSAL Courses must be submitted by November 2, 2009, to make the next catalog (2010--2011) production DATE (CHANGE DATE EACH TIME REVISED):
Drug Discovery. Zainab Al Kharusi Office: 33-10
 Drug Discovery Zainab Al Kharusi Office: 33-10 Objectives: To understand the processes of Drug Development To be able to develop a plan for drug discovery Reference: Patrick G L. An Introduction to Medicinal
Drug Discovery Zainab Al Kharusi Office: 33-10 Objectives: To understand the processes of Drug Development To be able to develop a plan for drug discovery Reference: Patrick G L. An Introduction to Medicinal
Lec.1 Chemistry Of Water
 Lec.1 Chemistry Of Water Biochemistry & Medicine Biochemistry can be defined as the science concerned with the chemical basis of life. Biochemistry can be described as the science concerned with the chemical
Lec.1 Chemistry Of Water Biochemistry & Medicine Biochemistry can be defined as the science concerned with the chemical basis of life. Biochemistry can be described as the science concerned with the chemical
Bioengineering & Bioinformatics Summer Institute, Dept. Computational Biology, University of Pittsburgh, PGH, PA
 Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Patrick: An Introduction to Medicinal Chemistry 5e Chapter 01
 Questions Patrick: An Introduction to Medicinal Chemistry 5e 01) Which of the following molecules is a phospholipid? a. i b. ii c. iii d. iv 02) Which of the following statements is false regarding the
Questions Patrick: An Introduction to Medicinal Chemistry 5e 01) Which of the following molecules is a phospholipid? a. i b. ii c. iii d. iv 02) Which of the following statements is false regarding the
Docking. GBCB 5874: Problem Solving in GBCB
 Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Practice Hour Examination # 1-1
 CHEM 346 Organic Chemistry I Fall 2013 Practice Hour Examination # 1-1 Solutions Key Page 1 of 12 CHEM 346 Organic Chemistry I (for Majors) Instructor: Paul J. Bracher Practice Hour Examination # 1-1 Monday,
CHEM 346 Organic Chemistry I Fall 2013 Practice Hour Examination # 1-1 Solutions Key Page 1 of 12 CHEM 346 Organic Chemistry I (for Majors) Instructor: Paul J. Bracher Practice Hour Examination # 1-1 Monday,
Introduction to FBDD Fragment screening methods and library design
 Introduction to FBDD Fragment screening methods and library design Samantha Hughes, PhD Fragments 2013 RSC BMCS Workshop 3 rd March 2013 Copyright 2013 Galapagos NV Why fragment screening methods? Guess
Introduction to FBDD Fragment screening methods and library design Samantha Hughes, PhD Fragments 2013 RSC BMCS Workshop 3 rd March 2013 Copyright 2013 Galapagos NV Why fragment screening methods? Guess
An automated synthesis programme for drug discovery
 An automated synthesis programme for drug discovery Automation has revolutionised the way that organic synthesis is performed in the research laboratory. Dr James Harness, Bohdan Automation Inc Combinatorial
An automated synthesis programme for drug discovery Automation has revolutionised the way that organic synthesis is performed in the research laboratory. Dr James Harness, Bohdan Automation Inc Combinatorial
Experiment 11: NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY
 Experiment 11: NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY Purpose: This is an exercise to introduce the use of nuclear magnetic resonance spectroscopy, in conjunction with infrared spectroscopy, to determine
Experiment 11: NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY Purpose: This is an exercise to introduce the use of nuclear magnetic resonance spectroscopy, in conjunction with infrared spectroscopy, to determine
Water: The Solvent for Biochemical Reactions
 Chapter 2 Water: The Solvent for Biochemical Reactions 11 SUMMARY Section 2.1 Section 2.2 Section 2.3 Section 2.4 Water is a polar molecule, with a partial negative charge on the oxygen and partial positive
Chapter 2 Water: The Solvent for Biochemical Reactions 11 SUMMARY Section 2.1 Section 2.2 Section 2.3 Section 2.4 Water is a polar molecule, with a partial negative charge on the oxygen and partial positive
19.4 Physical Properties Key: hydrogen bond strength depends on acidity of the hydrogen and basicity of the N or O
 Chem 360 Jasperse Ch. 19 Notes. Amines 12 19.4 ysical Properties Key: hydrogen bond strength depends on acidity of the hydrogen and basicity of the N or 1. Water Solubility: All amines hydrogen-bond water
Chem 360 Jasperse Ch. 19 Notes. Amines 12 19.4 ysical Properties Key: hydrogen bond strength depends on acidity of the hydrogen and basicity of the N or 1. Water Solubility: All amines hydrogen-bond water
Chapter 8 Educational Goals
 Chapter 8 Educational Goals 1. Given a chemical equation, write the law of mass action. 2. Given the equilibrium constant (K eq ) for a reaction, predict whether the reactants or products are predominant.
Chapter 8 Educational Goals 1. Given a chemical equation, write the law of mass action. 2. Given the equilibrium constant (K eq ) for a reaction, predict whether the reactants or products are predominant.
Structure based drug design and LIE models for GPCRs
 Structure based drug design and LIE models for GPCRs Peter Kolb kolb@docking.org Shoichet Lab ACS 237 th National Meeting, March 24, 2009 p.1/26 [Acknowledgements] Brian Shoichet John Irwin Mike Keiser
Structure based drug design and LIE models for GPCRs Peter Kolb kolb@docking.org Shoichet Lab ACS 237 th National Meeting, March 24, 2009 p.1/26 [Acknowledgements] Brian Shoichet John Irwin Mike Keiser
Lab #6: CARBOXYLIC ACIDS LAB
 lab Lab #6: CARBOXYLIC ACIDS LAB Name PART I: Preparation of Carboxylic Acids (a) Oxidation of an Aldehyde by Oxygen from the Air: Benzaldehyde is an aromatic aldehyde with a familiar odor. On a clean,
lab Lab #6: CARBOXYLIC ACIDS LAB Name PART I: Preparation of Carboxylic Acids (a) Oxidation of an Aldehyde by Oxygen from the Air: Benzaldehyde is an aromatic aldehyde with a familiar odor. On a clean,
10. Amines (text )
 2009, Department of Chemistry, The University of Western Ontario 10.1 10. Amines (text 10.1 10.6) A. Structure and omenclature Amines are derivatives of ammonia (H 3 ), where one or more H atoms has been
2009, Department of Chemistry, The University of Western Ontario 10.1 10. Amines (text 10.1 10.6) A. Structure and omenclature Amines are derivatives of ammonia (H 3 ), where one or more H atoms has been
Chapter 2 Water: The Solvent for Biochemical Reactions
 Chapter 2 Water: The Solvent for Biochemical Reactions SUMMARY Section 2.1 Water is a polar molecule, with a partial negative charge on the oxygen and partial positive charges on the hydrogens. There are
Chapter 2 Water: The Solvent for Biochemical Reactions SUMMARY Section 2.1 Water is a polar molecule, with a partial negative charge on the oxygen and partial positive charges on the hydrogens. There are
21.1 Introduction Carboxylic Acids Nomenclature of Carboxylic Acids. Acids Structure and Properties of Carboxylic Acids.
 21.1 Introduction Carboxylic Acids Carboxylic acids are abundant in nature and in pharmaceuticals. 21.1 Introduction Carboxylic Acids The US produces over 2.5 million tons of acetic acid per year, which
21.1 Introduction Carboxylic Acids Carboxylic acids are abundant in nature and in pharmaceuticals. 21.1 Introduction Carboxylic Acids The US produces over 2.5 million tons of acetic acid per year, which
Chapter-2 (Page 22-37) Physical and Chemical Properties of Water
 Chapter-2 (Page 22-37) Physical and Chemical Properties of Water Introduction About 70% of the mass of the human body is water. Water is central to biochemistry for the following reasons: 1- Biological
Chapter-2 (Page 22-37) Physical and Chemical Properties of Water Introduction About 70% of the mass of the human body is water. Water is central to biochemistry for the following reasons: 1- Biological
MOLECULAR DRUG TARGETS
 MOLECULAR DRUG TARGETS LEARNING OUTCOMES At the end of this session student shall be able to: List different types of druggable targets Describe forces involved in drug-receptor interactions Describe theories
MOLECULAR DRUG TARGETS LEARNING OUTCOMES At the end of this session student shall be able to: List different types of druggable targets Describe forces involved in drug-receptor interactions Describe theories
CHEMISTRY 332 SUMMER 08 EXAM I June 26-28, 2008
 First Three Letters of Last Name NAME Network ID CHEMISTRY 332 SUMMER 08 EXAM I June 26-28, 2008 The following materials are permissible during the exam: molecular model kits, course notes (printed, electronic,
First Three Letters of Last Name NAME Network ID CHEMISTRY 332 SUMMER 08 EXAM I June 26-28, 2008 The following materials are permissible during the exam: molecular model kits, course notes (printed, electronic,
Analysis of a Large Structure/Biological Activity. Data Set Using Recursive Partitioning and. Simulated Annealing
 Analysis of a Large Structure/Biological Activity Data Set Using Recursive Partitioning and Simulated Annealing Student: Ke Zhang MBMA Committee: Dr. Charles E. Smith (Chair) Dr. Jacqueline M. Hughes-Oliver
Analysis of a Large Structure/Biological Activity Data Set Using Recursive Partitioning and Simulated Annealing Student: Ke Zhang MBMA Committee: Dr. Charles E. Smith (Chair) Dr. Jacqueline M. Hughes-Oliver
Lecture 3: ph and Buffers
 Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2017 Lecture 3: ph and Buffers Tuesday, 16 Jan. 2017 c David P. Goldenberg University of Utah goldenberg@biology.utah.edu A Special Measure
Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2017 Lecture 3: ph and Buffers Tuesday, 16 Jan. 2017 c David P. Goldenberg University of Utah goldenberg@biology.utah.edu A Special Measure
OXFORD H i g h e r E d u c a t i o n Oxford University Press, All rights reserved.
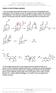 Patrick, An Introduction to dicinal hemistry 4e Answers to end-of-chapter questions 1) The mechanism below shows the release of one molecule of formaldehyde from methenamine. The mechanism can then be
Patrick, An Introduction to dicinal hemistry 4e Answers to end-of-chapter questions 1) The mechanism below shows the release of one molecule of formaldehyde from methenamine. The mechanism can then be
Chemistry 6/15/2015. Outline. Why study chemistry? Chemistry is the basis for studying much of biology.
 Chemistry Biology 105 Lecture 2 Reading: Chapter 2 (pages 20-29) Outline Why study chemistry??? Elements Atoms Periodic Table Electrons Bonding Bonds Covalent bonds Polarity Ionic bonds Hydrogen bonding
Chemistry Biology 105 Lecture 2 Reading: Chapter 2 (pages 20-29) Outline Why study chemistry??? Elements Atoms Periodic Table Electrons Bonding Bonds Covalent bonds Polarity Ionic bonds Hydrogen bonding
BSc and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes Marking
Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes Marking
Principles of Drug Design
 (16:663:502) Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu For more current information, please check WebCT at https://webct.rutgers.edu
(16:663:502) Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu For more current information, please check WebCT at https://webct.rutgers.edu
Reaxys Medicinal Chemistry Fact Sheet
 R&D SOLUTIONS FOR PHARMA & LIFE SCIENCES Reaxys Medicinal Chemistry Fact Sheet Essential data for lead identification and optimization Reaxys Medicinal Chemistry empowers early discovery in drug development
R&D SOLUTIONS FOR PHARMA & LIFE SCIENCES Reaxys Medicinal Chemistry Fact Sheet Essential data for lead identification and optimization Reaxys Medicinal Chemistry empowers early discovery in drug development
21.1 Introduction Carboxylic Acids
 21.1 Introduction Carboxylic Acids Carboxylic acids are abundant in nature and in pharmaceuticals. Klein, Organic Chemistry 1e 21-1 The US produces over 2.5 million tons of acetic acid per year, which
21.1 Introduction Carboxylic Acids Carboxylic acids are abundant in nature and in pharmaceuticals. Klein, Organic Chemistry 1e 21-1 The US produces over 2.5 million tons of acetic acid per year, which
10/16/17 ACIDS AND BASES, DEFINED WATER IS AMPHOTERIC OUTLINE. 9.1 Properties of Acids and Bases. 9.2 ph. 9.3 Buffers
 ACIDS AND BASES, DEFINED A hydrogen atom contains a proton and an electron, thus a hydrogen ion (H + ) is a proton: Acids: Proton (H + ) transfer between molecules is the basis of acid/base chemistry Ø
ACIDS AND BASES, DEFINED A hydrogen atom contains a proton and an electron, thus a hydrogen ion (H + ) is a proton: Acids: Proton (H + ) transfer between molecules is the basis of acid/base chemistry Ø
16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit. Lecture 2 (9/11/17)
 16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit By Anthony Quintano - https://www.flickr.com/photos/quintanomedia/15071865580, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=38538291
16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit By Anthony Quintano - https://www.flickr.com/photos/quintanomedia/15071865580, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=38538291
Plan. Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics.
 Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
Chem 460 Laboratory Fall 2008 Experiment 3: Investigating Fumarase: ph Profile, Stereospecificity and Thermodynamics of Reaction
 1 Chem 460 Laboratory Fall 2008 Experiment 3: Investigating Fumarase: ph Profile, Stereospecificity and Thermodynamics of Reaction Before Lab Week 1 -- ph Profile for Fumarase Read Box 11-1 (page 323)
1 Chem 460 Laboratory Fall 2008 Experiment 3: Investigating Fumarase: ph Profile, Stereospecificity and Thermodynamics of Reaction Before Lab Week 1 -- ph Profile for Fumarase Read Box 11-1 (page 323)
QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression
 APPLICATION NOTE QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression GAINING EFFICIENCY IN QUANTITATIVE STRUCTURE ACTIVITY RELATIONSHIPS ErbB1 kinase is the cell-surface receptor
APPLICATION NOTE QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression GAINING EFFICIENCY IN QUANTITATIVE STRUCTURE ACTIVITY RELATIONSHIPS ErbB1 kinase is the cell-surface receptor
2. WATER : THE SOLVENT FOR BIOCHEMICAL REACTIONS
 2. WATER : THE SOLVENT FOR BIOCHEMICAL REACTIONS 2.1 Water and Polarity Both geometry and properties of molecule determine polarity Electronegativity - The tendency of an atom to attract electrons to itself
2. WATER : THE SOLVENT FOR BIOCHEMICAL REACTIONS 2.1 Water and Polarity Both geometry and properties of molecule determine polarity Electronegativity - The tendency of an atom to attract electrons to itself
CHEMISTRY OF THE HUMAN BODY
 CHEMISTRY OF THE HUMAN BODY (Sample Questions Key) WUCT 2018 The three questions below are meant to give a sense of the kinds of questions that might be asked on the exam in April 2018. The actual exam
CHEMISTRY OF THE HUMAN BODY (Sample Questions Key) WUCT 2018 The three questions below are meant to give a sense of the kinds of questions that might be asked on the exam in April 2018. The actual exam
Chapter 25: The Chemistry of Life: Organic and Biological Chemistry
 Chemistry: The Central Science Chapter 25: The Chemistry of Life: Organic and Biological Chemistry The study of carbon compounds constitutes a separate branch of chemistry known as organic chemistry The
Chemistry: The Central Science Chapter 25: The Chemistry of Life: Organic and Biological Chemistry The study of carbon compounds constitutes a separate branch of chemistry known as organic chemistry The
How to make pyridines: the Hantzsch pyridine synthesis
 ow to make pyridines: the antzsch pyridine synthesis 1191 Zinc in acetic acid (Chapter 24) reduces the oxime to the amine and we can start the synthesis by doing the conjugate addition and then reducing
ow to make pyridines: the antzsch pyridine synthesis 1191 Zinc in acetic acid (Chapter 24) reduces the oxime to the amine and we can start the synthesis by doing the conjugate addition and then reducing
A primer on pharmacology pharmacodynamics
 A primer on pharmacology pharmacodynamics Drug binding & effect Universidade do Algarve Faro 2017 by Ferdi Engels, Ph.D. 1 Pharmacodynamics Relation with pharmacokinetics? dosage plasma concentration site
A primer on pharmacology pharmacodynamics Drug binding & effect Universidade do Algarve Faro 2017 by Ferdi Engels, Ph.D. 1 Pharmacodynamics Relation with pharmacokinetics? dosage plasma concentration site
HW #10: 10.38, 10.40, 10.46, 10.52, 10.58, 10.66, 10.68, 10.74, 10.78, 10.84, 10.88, 10.90, ,
 Chemistry 121 Lectures 20 & 21: Brønstead-Lowry Acid/Base Theory Revisited; Acid & Base Strength - Acids & Bases in Aqueous Solution; Acid Dissociation Constants and the Autoionization of Water; ph or
Chemistry 121 Lectures 20 & 21: Brønstead-Lowry Acid/Base Theory Revisited; Acid & Base Strength - Acids & Bases in Aqueous Solution; Acid Dissociation Constants and the Autoionization of Water; ph or
ALE 4. Effect of Temperature and Catalysts on the Rate of a Chemical Reaction
 Name Chem 163 Section: Team Number: ALE 4. Effect of Temperature and Catalysts on the Rate of a Chemical Reaction (Reference: 16.5 16.6 & 16.8 Silberberg 5 th edition) Why do reaction rates increase as
Name Chem 163 Section: Team Number: ALE 4. Effect of Temperature and Catalysts on the Rate of a Chemical Reaction (Reference: 16.5 16.6 & 16.8 Silberberg 5 th edition) Why do reaction rates increase as
Structure-Based Drug Discovery An Overview
 Structure-Based Drug Discovery An Overview Edited by Roderick E. Hubbard University of York, Heslington, York, UK and Vernalis (R&D) Ltd, Abington, Cambridge, UK RSC Publishing Contents Chapter 1 3D Structure
Structure-Based Drug Discovery An Overview Edited by Roderick E. Hubbard University of York, Heslington, York, UK and Vernalis (R&D) Ltd, Abington, Cambridge, UK RSC Publishing Contents Chapter 1 3D Structure
DOCKING TUTORIAL. A. The docking Workflow
 2 nd Strasbourg Summer School on Chemoinformatics VVF Obernai, France, 20-24 June 2010 E. Kellenberger DOCKING TUTORIAL A. The docking Workflow 1. Ligand preparation It consists in the standardization
2 nd Strasbourg Summer School on Chemoinformatics VVF Obernai, France, 20-24 June 2010 E. Kellenberger DOCKING TUTORIAL A. The docking Workflow 1. Ligand preparation It consists in the standardization
Chemical and Physical Properties of Organic Molecules
 Chemical and Physical Properties of Organic Molecules I.Elements A. Chemical symbols: C H O P S N C=carbon, H=hydrogen, O=oxygen, P=phosphorus, S=sulfur, N=nitrogen B. Top 3 Earth s surface = O, Si, Al
Chemical and Physical Properties of Organic Molecules I.Elements A. Chemical symbols: C H O P S N C=carbon, H=hydrogen, O=oxygen, P=phosphorus, S=sulfur, N=nitrogen B. Top 3 Earth s surface = O, Si, Al
CHAPTER 2. Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules
 CHAPTER 2 Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules 2-1 Kinetics and Thermodynamics of Simple Chemical Processes Chemical thermodynamics: Is concerned with the extent that
CHAPTER 2 Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules 2-1 Kinetics and Thermodynamics of Simple Chemical Processes Chemical thermodynamics: Is concerned with the extent that
FRAUNHOFER IME SCREENINGPORT
 FRAUNHOFER IME SCREENINGPORT Design of screening projects General remarks Introduction Screening is done to identify new chemical substances against molecular mechanisms of a disease It is a question of
FRAUNHOFER IME SCREENINGPORT Design of screening projects General remarks Introduction Screening is done to identify new chemical substances against molecular mechanisms of a disease It is a question of
Using NMR and IR Spectroscopy to Determine Structures Dr. Carl Hoeger, UCSD
 Using NMR and IR Spectroscopy to Determine Structures Dr. Carl Hoeger, UCSD The following guidelines should be helpful in assigning a structure from NMR (both PMR and CMR) and IR data. At the end of this
Using NMR and IR Spectroscopy to Determine Structures Dr. Carl Hoeger, UCSD The following guidelines should be helpful in assigning a structure from NMR (both PMR and CMR) and IR data. At the end of this
CHEM 347 Organic Chemistry II (for Majors) Instructor: Paul J. Bracher. Quiz # 4. Due in Monsanto Hall 103 by: Friday, April 4 th, 2014, 7:00 p.m.
 CHEM 347 Quiz # 4 Spring 2014 Page 1 of 9 CHEM 347 Organic Chemistry II (for Majors) Instructor: Paul J. Bracher Quiz # 4 Due in Monsanto Hall 103 by: Friday, April 4 th, 2014, 7:00 p.m. Student Name (Printed)
CHEM 347 Quiz # 4 Spring 2014 Page 1 of 9 CHEM 347 Organic Chemistry II (for Majors) Instructor: Paul J. Bracher Quiz # 4 Due in Monsanto Hall 103 by: Friday, April 4 th, 2014, 7:00 p.m. Student Name (Printed)
Overview: The Molecule That Supports All of Life
 Overview: The Molecule That Supports All of Life Water is the biological medium on Earth All living organisms require water more than any other substance Most cells are surrounded by water, and cells themselves
Overview: The Molecule That Supports All of Life Water is the biological medium on Earth All living organisms require water more than any other substance Most cells are surrounded by water, and cells themselves
Unlocking the potential of your drug discovery programme
 Unlocking the potential of your drug discovery programme Innovative screening The leading fragment screening platform with MicroScale Thermophoresis at its core Domainex expertise High quality results
Unlocking the potential of your drug discovery programme Innovative screening The leading fragment screening platform with MicroScale Thermophoresis at its core Domainex expertise High quality results
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability
 Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
CHM Salicylic Acid Properties (r16) 1/11
 CHM 111 - Salicylic Acid Properties (r16) 1/11 Purpose In this lab, you will perform several tests to attempt to confirm the identity and assess the purity of the substance you synthesized in last week's
CHM 111 - Salicylic Acid Properties (r16) 1/11 Purpose In this lab, you will perform several tests to attempt to confirm the identity and assess the purity of the substance you synthesized in last week's
