Learning Bayesian Networks Does Not Have to Be NP-Hard
|
|
|
- Sydney Mitchell
- 6 years ago
- Views:
Transcription
1 Learning Bayesian Networks Does Not Have to Be NP-Hard Norbert Dojer Institute of Informatics, Warsaw University, Banacha, Warszawa, Poland Abstract. We propose an algorithm for learning an optimal Bayesian network from data. Our method is addressed to biological applications, where usually datasets are small but sets of random variables are large. Moreover we assume that there is no need to examine the acyclicity of the graph. We provide polynomial bounds (with respect to the number of random variables) for time complexity of our algorithm for two generally used scoring criteria: Minimal Description Length and Bayesian-Dirichlet equivalence. 1 Introduction The framework of Bayesian networks is widely used in computational molecular biology. In particular, it appears attractive in the field of inferring a gene regulatory network structure from microarray expression data. Researchers dealing with Bayesian networks have generally accepted that without restrictive assumptions, learning Bayesian networks from data is NPhard with respect to the number of network vertices. This belief originates from a number of discouraging complexity results, which emerged over the last few years. Chickering [1] shows that learning an optimal network structure is NPhard for the BDe scoring criterion with an arbitrary prior distribution, even when each node has at most two parents and the dataset is trivial. In this approach a reduction of a known NP-complete problem is based on constructing a complicated prior distribution for the BDe score. Chickering et al. [] show that learning an optimal network structure is NP-hard for large datasets, even when each node has at most three parents and a scoring criterion favors the simplest model able to represent the distribution underlying the dataset exactly (as most of scores used in practice do). By a large dataset the authors mean the one which reflects the underlying distribution exactly. Moreover, this distribution is assumed to be directly accessible, so the complexity of scanning the dataset does not influence the result. In other papers [3,4], NP-hardness of the problem of learning Bayesian networks from data with some constraints on the structure of a network is shown. On the other hand, the known algorithms allow to learn the structure of optimal networks having up to 0-40 vertices [5]. Consequently, a large amount R. Královic and P. Urzyczyn (Eds.): MFCS 006, LNCS 416, pp , 006. c Springer-Verlag Berlin Heidelberg 006
2 306 N. Dojer of work has been dedicated to heuristic search techniques of identifying good models [6,7]. In the present paper we propose a new algorithm for learning an optimal network structure. Our method is addressed to the case when a dataset is small and there is no need to examine the acyclicity of the graph. The first condition does not bother in biological applications, because expenses of microarray experiments restrict the amount of expression data. The second assumption is satisfied in many cases, e.g. when working with dynamic Bayesian networks or when the sets of regulators and regulatees are disjoint. We investigate the worst-case time complexity of our algorithm for generally used scoring criteria. We show that the algorithm works in polynomial time for both MDL and BDe scores. Experiments with real biological data show that, even for large floral genomes, the algorithm for the MDL score works in a reasonable time. Algorithm A Bayesian network (BN) N is a representation of a joint distribution of a set of discrete random variables X = {X 1,...,X n }. The representation consists of two components: a directed acyclic graph G =(X, E) encoding conditional (in-)dependencies a family θ of conditional distributions P (X i Pa i ), where The joint distribution of X is given by Pa i = {Y X (Y,X i ) E} P (X) = n P (X i Pa i ) (1) i=1 The problem of learning a BN is understood as follows: given a multiset of X-instances D = {x 1,...,x N } find a network graph G that best matches D. The notion of a good match is formalized by means of a scoring function S(G : D) having positive values and minimized for the best matching network. Thus the point is to find a directed acyclic graph G with the set of vertices X minimizing S(G : D). In the present paper we consider the case when there is no need to examine the acyclicity of the graph, for example: When edges must be directed according to a given partial order, in particular when the sets of potential regulators and regulatees are disjoint. When dealing with dynamic Bayesian networks. A dynamic BN describes stochastic evolution of a set of random variables over discretized time. Therefore conditional distributions refer to random variables in neighboring time points. The acyclicity constraint is relaxed, because the unrolled graph
3 Learning Bayesian Networks Does Not Have to Be NP-Hard 307 (with a copy of each variable in each time point) is always acyclic (see [7,8] for more details). The following considerations apply to dynamic BNs as well. In the sequel we consider some assumptions on the form of a scoring function. The first one states that S(G : D) decomposes into a sum over the set of random variables of local scores, depending on the values of a variable and its parents in the graph only. Assumption 1 (additivity). S(G : D) = n i=1 s(x i, Pa i : D {Xi} Pa i ),where D Y denotes the restriction of D to the values of the members of Y X. When there is no need to examine the acyclicity of the graph, this assumption allows to compute the parents set of each variable independently. Thus the point is to find Pa i minimizing s(x i, Pa i : D {Xi} Pa i )foreachi. Let us fix a dataset D and a random variable X. WedenotebyX the set of potential parents of X (possibly smaller than X due to given constraints on the structure of the network). To simplify the notation we continue to write s(pa) for s(x, Pa : D {X} Pa ). The following assumption expresses the fact that scoring functions decompose into components: g penalizing the complexity of a network and d evaluating the possibility of explaining data by a network. Assumption (splitting). s(pa) =g(pa) +d(pa) for some functions g, d : P(X) R + satisfying Pa Pa = g(pa) g(pa ). This assumption is used in the following algorithm to avoid considering networks with inadequately large component g. Algorithm Pa :=. for each P X chosen according to g(p) (a) if s(p) <s(pa) thenpa := P (b) if g(p) s(pa) then return Pa; stop In the above algorithm choosing according to g(p) means choosing increasingly with respect to the value of the component g of the local score. Theorem 1. Suppose that the scoring function satisfies Assumptions 1-. Then Algorithm 1 applied to each random variable finds an optimal network. Proof. The theorem follows from the fact that all the potential parents sets P omitted by the algorithm satisfy s(p) g(p) s(pa), where Pa is the returned set.
4 308 N. Dojer A disadvantage of the above algorithm is that finding a proper subset P X involves computing g(p ) for all -successors P of previously chosen subsets. It may be avoided when a further assumption is imposed. Assumption 3 (uniformity). Pa = Pa = g(pa) =g(pa ). The above assumption suggests the notation ĝ( Pa ) = g(pa). The following algorithm uses the uniformity of g to reduce the number of computations of the component g. Algorithm. 1. Pa :=. for p =1ton (a) if ĝ(p) s(pa) then return Pa; stop (b) P = argmin {Y X : Y =p}s(y) (c) if s(p) <s(pa) thenpa := P Theorem. Suppose that the scoring function satisfies Assumptions 1-3. Then Algorithm applied to each random variable finds an optimal network. Proof. The theorem follows from the fact that all the potential parents sets P omitted by the algorithm satisfy s(p) ĝ( P ) s(pa), where Pa is the returned set. The following sections are devoted to two generally used scoring criteria: Minimal Description Length and Bayesian-Dirichlet equivalence. We consider possible decompositions of the local scores and analyze the computational cost of the above algorithms. 3 Minimal Description Length The Minimal Description Length (MDL) scoring criterion originates from information theory [9]. A network N is viewed here as a model of compression of a dataset D. The optimal model minimizes the total length of the description, i.e. the sum of the description length of the model and of the compressed data. Let us fix a dataset D = {x 1,...,x N } and a random variable X. Recall the decomposition s(pa) = g(pa) + d(pa) of the local score for X. IntheMDL score g(pa) stands for the length of the description of the local part of the network (i.e. the edges ingoing to X and the conditional distribution P (X Pa)) and d(pa) is the length of the compressed version of X-values in D. Let k Y denote the cardinality of the set V Y of possible values of the random variable Y X. Thuswehave g(pa) = Pa log n + log N (k X 1) k Y Y Pa
5 Learning Bayesian Networks Does Not Have to Be NP-Hard 309 where log N is the number of bits we use for each numeric parameter of the conditional distribution. This formula satisfies Assumption but fails to satisfy Assumption 3. Therefore Algorithm 1 can be used to learn an optimal network, but Algorithm cannot. However, for many applications we may assume that all the random variables attain values from the same set V of cardinality k. In this case we obtain the formula g(pa) = Pa log n + log N (k 1)k Pa which satisfies Assumption 3. For simplicity, we continue to work under this assumption. The general case may be handled in much the same way. Compression with respect to the network model is understood as follows: when encoding the X-values, the values of Pa-instances are assumed to be known. Thus the optimal encoding length is given by d(pa) =N H(X Pa) where H(X Pa) = P (v, v)logp (v v) is the conditional entropy Pa of X given Pa (the distributions are estimated from D). Since all the assumptions from the previous section are satisfied, Algorithm may be applied to learn the optimal network. Let us turn to the analysis of its complexity. Theorem 3. The worst-case time complexity of Algorithm for the MDL score is O(n log k N N log k N). Proof. The proof consists in finding a number p satisfying ĝ(p) s( ). Given v V,wedenotebyN v the number of samples in D with X = v. By Jensen s Theorem we have d( ) =N H(X ) =N N v N log N N log 1=Nlog k N v Hence s( ) N log k +(k 1) log N and it suffices to find p satisfying p log n + k p (k 1) log N N log k +(k 1) log N It is easily seen that the above assertion holds for p log k N whenever N>5. Therefore we do not have to consider subsets with more than log k N parent variables. Since there are O(n p ) subsets with at most p parents and a subset with p elements may be examined in pn steps, the total complexity of the algorithm is O(n log k N N log k N). 4 Bayesian-Dirichlet Equivalence The Bayesian-Dirichlet equivalence (BDe) scoring criterion originates from Bayesian statistics [10]. Given a dataset D the optimal network structure G maximizes the posterior conditional probability P (G D). We have
6 310 N. Dojer P (G D) P (G)P (D G) =P (G) P (D G,θ)P (θ G)dθ where P (G) and P (θ G) are prior probability distributions on graph structures and conditional distributions parameters, respectively, and P (D G,θ)isevaluated due to (1). Heckerman et al. [6], following Cooper and Herskovits [10], identified a set of independence assumptions making possible decomposition of the integral in the above formula into a product over X. Under this condition, together with a similar one regarding decomposition of P (G), the scoring criterion S(G : D) = log P (G) log P (D G) obtained by taking log of the above term satisfies Assumption 1. Moreover, the decomposition s(pa) = g(pa)+ d(pa) of the local scores appears as well, with the components g and d derived from log P (G) and log P (D G), respectively. The distribution P ((X, E)) α E with a penalty parameter 0 <α<1in general is used as a prior over the network structures. This choice results in the function g( Pa ) = Pa log α 1 satisfying Assumptions and 3. However, it should be noticed that there are also used priors which satisfy neither Assumption nor 3, e.g. P (G) α Δ(G,G0),whereΔ(G, G 0 )isthecardinality of the symmetric difference between the sets of edges in G andintheprior network G 0. The Dirichlet distribution is generally used as a prior over the conditional distributions parameters. It yields d(pa) =log Γ ( (H v,v + N v,v )) Γ ( Γ (H v,v ) H v,v) Γ (H v,v + N v,v ) Pa where Γ is the Gamma function, N v,v denotes the number of samples in D with X = v and Pa = v, andh v,v is the corresponding hyperparameter of the Dirichlet distribution. Setting all the hyperparameters to 1 yields d(pa) =log Pa = Pa (k 1+ N v,v)! 1 = (k 1)! N v,v! ( log(k 1+ N v,v )! log(k 1)! ) log N v,v! where k = V. For simplicity, we continue to work under this assumption. The general case may be handled in a similar way. The following result allows to refine the decomposition of the local score into the sum of the components g and d.
7 Learning Bayesian Networks Does Not Have to Be NP-Hard 311 Proposition 1. Define d min = (log(k 1+N v)! log(k 1)! log N v!), where N v denotes the number of samples in D with X = v. Thend(Pa) d min for each Pa X. Proof. Fix Pa X. Givenv V Pa,wedenotebyN v the number of samples in D with Pa = v. Wehave d(pa) = ( log(k 1+N v )! log(k 1)! ) log N v,v! = Pa = k 1+N v log i N v,v log i Pa i=1 k 1+N v,v N v,v log i log i = N v,v log k 1+i = i Pa = = N v,v Pa i=1 i=1 i=1 log( k 1 i N v log k 1+i i = = +1) ( k 1+Nv Pa i=1 N v log( k 1 i i=1 +1)= ) N v log i log i = i=1 (log(k 1+N v )! log(k 1)! log N v!) = d min The first inequality follows from the fact that given N v the sum k 1+N v,v log i maximizes when N v = N v,v for some v V. Similarly, the second inequality holds, because given N v the sum N v,v Pa i=1 log( k 1 i +1) minimizes when N v = N v,v for some v V Pa. By the above proposition, the decomposition of the local score given by s(pa) = g (Pa) +d (Pa) with the components g (Pa) =g(pa) +d min and d (Pa) = d(pa) d min satisfies all the assumptions required by Algorithm. Let us turn to the analysis of its complexity. Theorem 4. The worst-case time complexity of Algorithm for the BDe score with the decomposition of the local score given by s(pa) =g (Pa) +d (Pa) is O(n N log α 1 k N log α 1 k).
8 31 N. Dojer Proof. The proof consists in finding a number p satisfying ĝ (p) s( ). We have d ( ) =d( ) d min = ( (k 1+N)! = log ) log N v! (k 1)! = (log(k 1+N)! log(k 1)!) k 1+N =loge = log i k k 1+N k 1+N ln i k k 1+ N k log i k 1+ N k ( ( k+n k 1+ N log e ln xdx k k ( log (k 1+N ) v)! log N v! = (k 1)! (log(k 1+N v )! log(k 1)!) = k 1+N v log i log i +(N k N k )log(k + N k ) = k 1 ln i +( N k N k )ln(k + N k ) k )) k 1+ N k ln xdx+ ln xdx = k 1+ N k ( =loge (k + N)(ln(k + N) 1) k(ln k 1) ( k (k 1+ N ) ) k )(ln(k 1+N k ) 1) (k 1)(ln(k 1) 1) = k + N = N log k + N log k k + N +log (1 + N k )k (1 + N N log k k k )k k The first inequality follows from the fact that the sum log N v! minimizes when the values of N v are uniformly distributed over v. The last inequality is due to the fact that k, and consequently k k k. Thus s( ) N log k + d min, so we need p satisfying p log α 1 N log k, i.e. p N log α 1 k. Therefore we do not have to consider subsets with more than N log α 1 k parent variables. Since there are O(n p )subsetswithatmostpparents and a subset with p elements may be examined in pn steps, the total complexity of the algorithm is O(n N log α 1 k N log α 1 k). 5 Application to Biological Data Microarray experiments measure simultaneously expression levels of thousands of genes. Learning Bayesian networks from microarray data aims at discovering gene regulatory dependencies.
9 Learning Bayesian Networks Does Not Have to Be NP-Hard 313 We tested the time complexity of learning an optimal dynamic BN on microarray experiments data of Arabidopsis thaliana ( genes). There were 0 samples of time series data discretized into 3 expression levels. We ran our program on a single Pentium 4 CPU.8 GHz. The computation time was 48 hours for the MDL score and 170 hours for the BDe score with a parameter log α 1 = 5. The program failed to terminate in a reasonable time for the BDe score with a parameter log α 1 = 3, i.e α Conclusion The aim of this work was to provide an effective algorithm for learning optimal Bayesian network from data, applicable to microarray expression measurements. We have proposed a method addressed to the case when a dataset is small and there is no need to examine the acyclicity of the graph. We have provided polynomial bounds (with respect to the number of random variables) for time complexity of our algorithm for two generally used scoring criteria: Minimal Description Length and Bayesian-Dirichlet equivalence. An application to real biological data shows that our algorithm works with large floral genomes in a reasonable time for the MDL score but fails to terminate for the BDe score with a sensible value of the penalty parameter. Acknowledgements The author is grateful to Jerzy Tiuryn for comments on previous versions of this work and useful discussions related to the topic. I also thank Bartek Wilczyński for implementing and running our learning algorithm. This research was founded by Polish State Committee for Scientific Research under grant no. 3T11F018. References 1. Chickering, D.M.: Learning Bayesian networks is NP-complete. In Fisher, D., Lenz, H.J., eds.: Learning from Data: Artificial Inteligence and Statistics V. Springer- Verlag (1996). Chickering, D.M., Heckerman, D., Meek, C.: Large-sample learning of Bayesian networks is NP-hard. Journal of Machine Learning Research 5 (004) Dasgupta, S.: Learning polytrees. In: Proceedings of the 15th Annual Conference on Uncertainty in Artificial Intelligence (UAI-99), San Francisco, CA, Morgan Kaufmann Publishers (1999) Meek, C.: Finding a path is harder than finding a tree. J. Artificial Intelligence Res. 15 (001) (electronic) 5. Ott, S., Imoto, S., Miyano, S.: Finding optimal models for small gene networks. Pac. Symp. Biocomput. (004) Heckerman, D., Geiger, D., Chickering, D.M.: Learning Bayesian networks: The combination of knowledge and statistical data. Machine Learning 0(3) (1995)
10 314 N. Dojer 7. Neapolitan, R.E.: Learning Bayesian Networks. Prentice Hall (003) 8. Friedman, N., Murphy, K., Russell, S.: Learning the structure of dynamic probabilistic networks. In Cooper, G.F., Moral, S., eds.: Proceedings of the Fourteenth Conference on Uncertainty in Artificial Inteligence. (1998) Lam, W., Bacchus, F.: Learning Bayesian belief networks: An approach based on the MDL principle. Computational Intelligence 10(3) (1994) Cooper, G.F., Herskovits, E.: A Bayesian method for the induction of probabilistic networks from data. Machine Learning 9 (199)
Learning in Bayesian Networks
 Learning in Bayesian Networks Florian Markowetz Max-Planck-Institute for Molecular Genetics Computational Molecular Biology Berlin Berlin: 20.06.2002 1 Overview 1. Bayesian Networks Stochastic Networks
Learning in Bayesian Networks Florian Markowetz Max-Planck-Institute for Molecular Genetics Computational Molecular Biology Berlin Berlin: 20.06.2002 1 Overview 1. Bayesian Networks Stochastic Networks
A Decision Theoretic View on Choosing Heuristics for Discovery of Graphical Models
 A Decision Theoretic View on Choosing Heuristics for Discovery of Graphical Models Y. Xiang University of Guelph, Canada Abstract Discovery of graphical models is NP-hard in general, which justifies using
A Decision Theoretic View on Choosing Heuristics for Discovery of Graphical Models Y. Xiang University of Guelph, Canada Abstract Discovery of graphical models is NP-hard in general, which justifies using
Model Complexity of Pseudo-independent Models
 Model Complexity of Pseudo-independent Models Jae-Hyuck Lee and Yang Xiang Department of Computing and Information Science University of Guelph, Guelph, Canada {jaehyuck, yxiang}@cis.uoguelph,ca Abstract
Model Complexity of Pseudo-independent Models Jae-Hyuck Lee and Yang Xiang Department of Computing and Information Science University of Guelph, Guelph, Canada {jaehyuck, yxiang}@cis.uoguelph,ca Abstract
Branch and Bound for Regular Bayesian Network Structure Learning
 Branch and Bound for Regular Bayesian Network Structure Learning Joe Suzuki and Jun Kawahara Osaka University, Japan. j-suzuki@sigmath.es.osaka-u.ac.jp Nara Institute of Science and Technology, Japan.
Branch and Bound for Regular Bayesian Network Structure Learning Joe Suzuki and Jun Kawahara Osaka University, Japan. j-suzuki@sigmath.es.osaka-u.ac.jp Nara Institute of Science and Technology, Japan.
Introduction to Probabilistic Graphical Models
 Introduction to Probabilistic Graphical Models Kyu-Baek Hwang and Byoung-Tak Zhang Biointelligence Lab School of Computer Science and Engineering Seoul National University Seoul 151-742 Korea E-mail: kbhwang@bi.snu.ac.kr
Introduction to Probabilistic Graphical Models Kyu-Baek Hwang and Byoung-Tak Zhang Biointelligence Lab School of Computer Science and Engineering Seoul National University Seoul 151-742 Korea E-mail: kbhwang@bi.snu.ac.kr
Scoring functions for learning Bayesian networks
 Tópicos Avançados p. 1/48 Scoring functions for learning Bayesian networks Alexandra M. Carvalho Tópicos Avançados p. 2/48 Plan Learning Bayesian networks Scoring functions for learning Bayesian networks:
Tópicos Avançados p. 1/48 Scoring functions for learning Bayesian networks Alexandra M. Carvalho Tópicos Avançados p. 2/48 Plan Learning Bayesian networks Scoring functions for learning Bayesian networks:
Discovery of Pseudo-Independent Models from Data
 Discovery of Pseudo-Independent Models from Data Yang Xiang, University of Guelph, Canada June 4, 2004 INTRODUCTION Graphical models such as Bayesian networks (BNs) and decomposable Markov networks (DMNs)
Discovery of Pseudo-Independent Models from Data Yang Xiang, University of Guelph, Canada June 4, 2004 INTRODUCTION Graphical models such as Bayesian networks (BNs) and decomposable Markov networks (DMNs)
Applying Bayesian networks in the game of Minesweeper
 Applying Bayesian networks in the game of Minesweeper Marta Vomlelová Faculty of Mathematics and Physics Charles University in Prague http://kti.mff.cuni.cz/~marta/ Jiří Vomlel Institute of Information
Applying Bayesian networks in the game of Minesweeper Marta Vomlelová Faculty of Mathematics and Physics Charles University in Prague http://kti.mff.cuni.cz/~marta/ Jiří Vomlel Institute of Information
Exact model averaging with naive Bayesian classifiers
 Exact model averaging with naive Bayesian classifiers Denver Dash ddash@sispittedu Decision Systems Laboratory, Intelligent Systems Program, University of Pittsburgh, Pittsburgh, PA 15213 USA Gregory F
Exact model averaging with naive Bayesian classifiers Denver Dash ddash@sispittedu Decision Systems Laboratory, Intelligent Systems Program, University of Pittsburgh, Pittsburgh, PA 15213 USA Gregory F
The Monte Carlo Method: Bayesian Networks
 The Method: Bayesian Networks Dieter W. Heermann Methods 2009 Dieter W. Heermann ( Methods)The Method: Bayesian Networks 2009 1 / 18 Outline 1 Bayesian Networks 2 Gene Expression Data 3 Bayesian Networks
The Method: Bayesian Networks Dieter W. Heermann Methods 2009 Dieter W. Heermann ( Methods)The Method: Bayesian Networks 2009 1 / 18 Outline 1 Bayesian Networks 2 Gene Expression Data 3 Bayesian Networks
COMP538: Introduction to Bayesian Networks
 COMP538: Introduction to Bayesian Networks Lecture 9: Optimal Structure Learning Nevin L. Zhang lzhang@cse.ust.hk Department of Computer Science and Engineering Hong Kong University of Science and Technology
COMP538: Introduction to Bayesian Networks Lecture 9: Optimal Structure Learning Nevin L. Zhang lzhang@cse.ust.hk Department of Computer Science and Engineering Hong Kong University of Science and Technology
Lecture 5: Bayesian Network
 Lecture 5: Bayesian Network Topics of this lecture What is a Bayesian network? A simple example Formal definition of BN A slightly difficult example Learning of BN An example of learning Important topics
Lecture 5: Bayesian Network Topics of this lecture What is a Bayesian network? A simple example Formal definition of BN A slightly difficult example Learning of BN An example of learning Important topics
The GlobalMIT Toolkit. Learning Optimal Dynamic Bayesian Network
 The GlobalMIT Toolkit for Learning Optimal Dynamic Bayesian Network User Guide Rev. 1.1 Maintained by Vinh Nguyen c 2010-2011 Vinh Xuan Nguyen All rights reserved Monash University, Victoria, Australia.
The GlobalMIT Toolkit for Learning Optimal Dynamic Bayesian Network User Guide Rev. 1.1 Maintained by Vinh Nguyen c 2010-2011 Vinh Xuan Nguyen All rights reserved Monash University, Victoria, Australia.
6.867 Machine learning, lecture 23 (Jaakkola)
 Lecture topics: Markov Random Fields Probabilistic inference Markov Random Fields We will briefly go over undirected graphical models or Markov Random Fields (MRFs) as they will be needed in the context
Lecture topics: Markov Random Fields Probabilistic inference Markov Random Fields We will briefly go over undirected graphical models or Markov Random Fields (MRFs) as they will be needed in the context
Exact distribution theory for belief net responses
 Exact distribution theory for belief net responses Peter M. Hooper Department of Mathematical and Statistical Sciences University of Alberta Edmonton, Canada, T6G 2G1 hooper@stat.ualberta.ca May 2, 2008
Exact distribution theory for belief net responses Peter M. Hooper Department of Mathematical and Statistical Sciences University of Alberta Edmonton, Canada, T6G 2G1 hooper@stat.ualberta.ca May 2, 2008
Abstract. Three Methods and Their Limitations. N-1 Experiments Suffice to Determine the Causal Relations Among N Variables
 N-1 Experiments Suffice to Determine the Causal Relations Among N Variables Frederick Eberhardt Clark Glymour 1 Richard Scheines Carnegie Mellon University Abstract By combining experimental interventions
N-1 Experiments Suffice to Determine the Causal Relations Among N Variables Frederick Eberhardt Clark Glymour 1 Richard Scheines Carnegie Mellon University Abstract By combining experimental interventions
Finite Mixture Model of Bounded Semi-naive Bayesian Networks Classifier
 Finite Mixture Model of Bounded Semi-naive Bayesian Networks Classifier Kaizhu Huang, Irwin King, and Michael R. Lyu Department of Computer Science and Engineering The Chinese University of Hong Kong Shatin,
Finite Mixture Model of Bounded Semi-naive Bayesian Networks Classifier Kaizhu Huang, Irwin King, and Michael R. Lyu Department of Computer Science and Engineering The Chinese University of Hong Kong Shatin,
Parent Assignment Is Hard for the MDL, AIC, and NML Costs
 Parent Assignment Is Hard for the MDL, AIC, and NML Costs Mikko Koivisto HIIT Basic Research Unit, Department of Computer Science Gustaf Hällströmin katu 2b, FIN-00014 University of Helsinki, Finland mikko.koivisto@cs.helsinki.fi
Parent Assignment Is Hard for the MDL, AIC, and NML Costs Mikko Koivisto HIIT Basic Research Unit, Department of Computer Science Gustaf Hällströmin katu 2b, FIN-00014 University of Helsinki, Finland mikko.koivisto@cs.helsinki.fi
Structure Learning: the good, the bad, the ugly
 Readings: K&F: 15.1, 15.2, 15.3, 15.4, 15.5 Structure Learning: the good, the bad, the ugly Graphical Models 10708 Carlos Guestrin Carnegie Mellon University September 29 th, 2006 1 Understanding the uniform
Readings: K&F: 15.1, 15.2, 15.3, 15.4, 15.5 Structure Learning: the good, the bad, the ugly Graphical Models 10708 Carlos Guestrin Carnegie Mellon University September 29 th, 2006 1 Understanding the uniform
Beyond Uniform Priors in Bayesian Network Structure Learning
 Beyond Uniform Priors in Bayesian Network Structure Learning (for Discrete Bayesian Networks) scutari@stats.ox.ac.uk Department of Statistics April 5, 2017 Bayesian Network Structure Learning Learning
Beyond Uniform Priors in Bayesian Network Structure Learning (for Discrete Bayesian Networks) scutari@stats.ox.ac.uk Department of Statistics April 5, 2017 Bayesian Network Structure Learning Learning
Bayesian Network Structure Learning using Factorized NML Universal Models
 Bayesian Network Structure Learning using Factorized NML Universal Models Teemu Roos, Tomi Silander, Petri Kontkanen, and Petri Myllymäki Complex Systems Computation Group, Helsinki Institute for Information
Bayesian Network Structure Learning using Factorized NML Universal Models Teemu Roos, Tomi Silander, Petri Kontkanen, and Petri Myllymäki Complex Systems Computation Group, Helsinki Institute for Information
An Empirical-Bayes Score for Discrete Bayesian Networks
 JMLR: Workshop and Conference Proceedings vol 52, 438-448, 2016 PGM 2016 An Empirical-Bayes Score for Discrete Bayesian Networks Marco Scutari Department of Statistics University of Oxford Oxford, United
JMLR: Workshop and Conference Proceedings vol 52, 438-448, 2016 PGM 2016 An Empirical-Bayes Score for Discrete Bayesian Networks Marco Scutari Department of Statistics University of Oxford Oxford, United
Belief Update in CLG Bayesian Networks With Lazy Propagation
 Belief Update in CLG Bayesian Networks With Lazy Propagation Anders L Madsen HUGIN Expert A/S Gasværksvej 5 9000 Aalborg, Denmark Anders.L.Madsen@hugin.com Abstract In recent years Bayesian networks (BNs)
Belief Update in CLG Bayesian Networks With Lazy Propagation Anders L Madsen HUGIN Expert A/S Gasværksvej 5 9000 Aalborg, Denmark Anders.L.Madsen@hugin.com Abstract In recent years Bayesian networks (BNs)
An Empirical-Bayes Score for Discrete Bayesian Networks
 An Empirical-Bayes Score for Discrete Bayesian Networks scutari@stats.ox.ac.uk Department of Statistics September 8, 2016 Bayesian Network Structure Learning Learning a BN B = (G, Θ) from a data set D
An Empirical-Bayes Score for Discrete Bayesian Networks scutari@stats.ox.ac.uk Department of Statistics September 8, 2016 Bayesian Network Structure Learning Learning a BN B = (G, Θ) from a data set D
Comparing Bayesian Networks and Structure Learning Algorithms
 Comparing Bayesian Networks and Structure Learning Algorithms (and other graphical models) marco.scutari@stat.unipd.it Department of Statistical Sciences October 20, 2009 Introduction Introduction Graphical
Comparing Bayesian Networks and Structure Learning Algorithms (and other graphical models) marco.scutari@stat.unipd.it Department of Statistical Sciences October 20, 2009 Introduction Introduction Graphical
Polynomial-Time Algorithm for Learning Optimal BFS-Consistent Dynamic Bayesian Networks
 entropy Article Polynomial-Time Algorithm for Learning Optimal BFS-Consistent Dynamic Bayesian Networks Margarida Sousa and Alexandra M. Carvalho * Instituto de Telecomunicações, Instituto Superior Técnico,
entropy Article Polynomial-Time Algorithm for Learning Optimal BFS-Consistent Dynamic Bayesian Networks Margarida Sousa and Alexandra M. Carvalho * Instituto de Telecomunicações, Instituto Superior Técnico,
Computational Genomics. Systems biology. Putting it together: Data integration using graphical models
 02-710 Computational Genomics Systems biology Putting it together: Data integration using graphical models High throughput data So far in this class we discussed several different types of high throughput
02-710 Computational Genomics Systems biology Putting it together: Data integration using graphical models High throughput data So far in this class we discussed several different types of high throughput
Score Metrics for Learning Bayesian Networks used as Fitness Function in a Genetic Algorithm
 Score Metrics for Learning Bayesian Networks used as Fitness Function in a Genetic Algorithm Edimilson B. dos Santos 1, Estevam R. Hruschka Jr. 2 and Nelson F. F. Ebecken 1 1 COPPE/UFRJ - Federal University
Score Metrics for Learning Bayesian Networks used as Fitness Function in a Genetic Algorithm Edimilson B. dos Santos 1, Estevam R. Hruschka Jr. 2 and Nelson F. F. Ebecken 1 1 COPPE/UFRJ - Federal University
Markovian Combination of Decomposable Model Structures: MCMoSt
 Markovian Combination 1/45 London Math Society Durham Symposium on Mathematical Aspects of Graphical Models. June 30 - July, 2008 Markovian Combination of Decomposable Model Structures: MCMoSt Sung-Ho
Markovian Combination 1/45 London Math Society Durham Symposium on Mathematical Aspects of Graphical Models. June 30 - July, 2008 Markovian Combination of Decomposable Model Structures: MCMoSt Sung-Ho
Learning Bayesian Networks: The Combination of Knowledge and Statistical Data
 Learning Bayesian Networks: The Combination of Knowledge and Statistical Data David Heckerman Dan Geiger Microsoft Research, Bldg 9S Redmond, WA 98052-6399 David M. Chickering heckerma@microsoft.com, dang@cs.technion.ac.il,
Learning Bayesian Networks: The Combination of Knowledge and Statistical Data David Heckerman Dan Geiger Microsoft Research, Bldg 9S Redmond, WA 98052-6399 David M. Chickering heckerma@microsoft.com, dang@cs.technion.ac.il,
2 : Directed GMs: Bayesian Networks
 10-708: Probabilistic Graphical Models 10-708, Spring 2017 2 : Directed GMs: Bayesian Networks Lecturer: Eric P. Xing Scribes: Jayanth Koushik, Hiroaki Hayashi, Christian Perez Topic: Directed GMs 1 Types
10-708: Probabilistic Graphical Models 10-708, Spring 2017 2 : Directed GMs: Bayesian Networks Lecturer: Eric P. Xing Scribes: Jayanth Koushik, Hiroaki Hayashi, Christian Perez Topic: Directed GMs 1 Types
Polynomial-time algorithm for learning optimal tree-augmented dynamic Bayesian networks
 Polynomial-time algorithm for learning optimal tree-augmented dynamic Bayesian networks José L. Monteiro IDMEC Instituto Superior Técnico Universidade de Lisboa jose.libano.monteiro@tecnico.ulisboa.pt
Polynomial-time algorithm for learning optimal tree-augmented dynamic Bayesian networks José L. Monteiro IDMEC Instituto Superior Técnico Universidade de Lisboa jose.libano.monteiro@tecnico.ulisboa.pt
Learning Causal Bayesian Networks from Observations and Experiments: A Decision Theoretic Approach
 Learning Causal Bayesian Networks from Observations and Experiments: A Decision Theoretic Approach Stijn Meganck 1, Philippe Leray 2, and Bernard Manderick 1 1 Vrije Universiteit Brussel, Pleinlaan 2,
Learning Causal Bayesian Networks from Observations and Experiments: A Decision Theoretic Approach Stijn Meganck 1, Philippe Leray 2, and Bernard Manderick 1 1 Vrije Universiteit Brussel, Pleinlaan 2,
Machine Learning Lecture 14
 Many slides adapted from B. Schiele, S. Roth, Z. Gharahmani Machine Learning Lecture 14 Undirected Graphical Models & Inference 23.06.2015 Bastian Leibe RWTH Aachen http://www.vision.rwth-aachen.de/ leibe@vision.rwth-aachen.de
Many slides adapted from B. Schiele, S. Roth, Z. Gharahmani Machine Learning Lecture 14 Undirected Graphical Models & Inference 23.06.2015 Bastian Leibe RWTH Aachen http://www.vision.rwth-aachen.de/ leibe@vision.rwth-aachen.de
Entropy-based pruning for learning Bayesian networks using BIC
 Entropy-based pruning for learning Bayesian networks using BIC Cassio P. de Campos a,b,, Mauro Scanagatta c, Giorgio Corani c, Marco Zaffalon c a Utrecht University, The Netherlands b Queen s University
Entropy-based pruning for learning Bayesian networks using BIC Cassio P. de Campos a,b,, Mauro Scanagatta c, Giorgio Corani c, Marco Zaffalon c a Utrecht University, The Netherlands b Queen s University
Inferring Protein-Signaling Networks II
 Inferring Protein-Signaling Networks II Lectures 15 Nov 16, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022
Inferring Protein-Signaling Networks II Lectures 15 Nov 16, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022
Entropy-based Pruning for Learning Bayesian Networks using BIC
 Entropy-based Pruning for Learning Bayesian Networks using BIC Cassio P. de Campos Queen s University Belfast, UK arxiv:1707.06194v1 [cs.ai] 19 Jul 2017 Mauro Scanagatta Giorgio Corani Marco Zaffalon Istituto
Entropy-based Pruning for Learning Bayesian Networks using BIC Cassio P. de Campos Queen s University Belfast, UK arxiv:1707.06194v1 [cs.ai] 19 Jul 2017 Mauro Scanagatta Giorgio Corani Marco Zaffalon Istituto
Gearing optimization
 Gearing optimization V.V. Lozin Abstract We consider an optimization problem that arises in machine-tool design. It deals with optimization of the structure of gearbox, which is normally represented by
Gearing optimization V.V. Lozin Abstract We consider an optimization problem that arises in machine-tool design. It deals with optimization of the structure of gearbox, which is normally represented by
Bayesian Network Learning and Applications in Bioinformatics. Xiaotong Lin
 Bayesian Network Learning and Applications in Bioinformatics By Xiaotong Lin Submitted to the Department of Electrical Engineering and Computer Science and the Faculty of the Graduate School of the University
Bayesian Network Learning and Applications in Bioinformatics By Xiaotong Lin Submitted to the Department of Electrical Engineering and Computer Science and the Faculty of the Graduate School of the University
Inferring Protein-Signaling Networks
 Inferring Protein-Signaling Networks Lectures 14 Nov 14, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022 1
Inferring Protein-Signaling Networks Lectures 14 Nov 14, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 12:00-1:20 Johnson Hall (JHN) 022 1
Lecture 8: December 17, 2003
 Computational Genomics Fall Semester, 2003 Lecture 8: December 17, 2003 Lecturer: Irit Gat-Viks Scribe: Tal Peled and David Burstein 8.1 Exploiting Independence Property 8.1.1 Introduction Our goal is
Computational Genomics Fall Semester, 2003 Lecture 8: December 17, 2003 Lecturer: Irit Gat-Viks Scribe: Tal Peled and David Burstein 8.1 Exploiting Independence Property 8.1.1 Introduction Our goal is
{ p if x = 1 1 p if x = 0
 Discrete random variables Probability mass function Given a discrete random variable X taking values in X = {v 1,..., v m }, its probability mass function P : X [0, 1] is defined as: P (v i ) = Pr[X =
Discrete random variables Probability mass function Given a discrete random variable X taking values in X = {v 1,..., v m }, its probability mass function P : X [0, 1] is defined as: P (v i ) = Pr[X =
Directed Graphical Models or Bayesian Networks
 Directed Graphical Models or Bayesian Networks Le Song Machine Learning II: Advanced Topics CSE 8803ML, Spring 2012 Bayesian Networks One of the most exciting recent advancements in statistical AI Compact
Directed Graphical Models or Bayesian Networks Le Song Machine Learning II: Advanced Topics CSE 8803ML, Spring 2012 Bayesian Networks One of the most exciting recent advancements in statistical AI Compact
Integer Programming for Bayesian Network Structure Learning
 Integer Programming for Bayesian Network Structure Learning James Cussens Helsinki, 2013-04-09 James Cussens IP for BNs Helsinki, 2013-04-09 1 / 20 Linear programming The Belgian diet problem Fat Sugar
Integer Programming for Bayesian Network Structure Learning James Cussens Helsinki, 2013-04-09 James Cussens IP for BNs Helsinki, 2013-04-09 1 / 20 Linear programming The Belgian diet problem Fat Sugar
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
Rapid Introduction to Machine Learning/ Deep Learning
 Rapid Introduction to Machine Learning/ Deep Learning Hyeong In Choi Seoul National University 1/32 Lecture 5a Bayesian network April 14, 2016 2/32 Table of contents 1 1. Objectives of Lecture 5a 2 2.Bayesian
Rapid Introduction to Machine Learning/ Deep Learning Hyeong In Choi Seoul National University 1/32 Lecture 5a Bayesian network April 14, 2016 2/32 Table of contents 1 1. Objectives of Lecture 5a 2 2.Bayesian
Structure Learning of Bayesian Networks using Constraints
 Cassio P. de Campos Dalle Molle Institute for Artificial Intelligence (IDSIA), Galleria 2, Manno 6928, Switzerland Zhi Zeng Qiang Ji Rensselaer Polytechnic Institute (RPI), 110 8th St., Troy NY 12180,
Cassio P. de Campos Dalle Molle Institute for Artificial Intelligence (IDSIA), Galleria 2, Manno 6928, Switzerland Zhi Zeng Qiang Ji Rensselaer Polytechnic Institute (RPI), 110 8th St., Troy NY 12180,
Probabilistic Graphical Models (I)
 Probabilistic Graphical Models (I) Hongxin Zhang zhx@cad.zju.edu.cn State Key Lab of CAD&CG, ZJU 2015-03-31 Probabilistic Graphical Models Modeling many real-world problems => a large number of random
Probabilistic Graphical Models (I) Hongxin Zhang zhx@cad.zju.edu.cn State Key Lab of CAD&CG, ZJU 2015-03-31 Probabilistic Graphical Models Modeling many real-world problems => a large number of random
Food delivered. Food obtained S 3
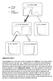 Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Bayesian Network Classifiers *
 Machine Learning, 29, 131 163 (1997) c 1997 Kluwer Academic Publishers. Manufactured in The Netherlands. Bayesian Network Classifiers * NIR FRIEDMAN Computer Science Division, 387 Soda Hall, University
Machine Learning, 29, 131 163 (1997) c 1997 Kluwer Academic Publishers. Manufactured in The Netherlands. Bayesian Network Classifiers * NIR FRIEDMAN Computer Science Division, 387 Soda Hall, University
Learning Bayesian Networks for Biomedical Data
 Learning Bayesian Networks for Biomedical Data Faming Liang (Texas A&M University ) Liang, F. and Zhang, J. (2009) Learning Bayesian Networks for Discrete Data. Computational Statistics and Data Analysis,
Learning Bayesian Networks for Biomedical Data Faming Liang (Texas A&M University ) Liang, F. and Zhang, J. (2009) Learning Bayesian Networks for Discrete Data. Computational Statistics and Data Analysis,
Induction of Decision Trees
 Induction of Decision Trees Peter Waiganjo Wagacha This notes are for ICS320 Foundations of Learning and Adaptive Systems Institute of Computer Science University of Nairobi PO Box 30197, 00200 Nairobi.
Induction of Decision Trees Peter Waiganjo Wagacha This notes are for ICS320 Foundations of Learning and Adaptive Systems Institute of Computer Science University of Nairobi PO Box 30197, 00200 Nairobi.
12 : Variational Inference I
 10-708: Probabilistic Graphical Models, Spring 2015 12 : Variational Inference I Lecturer: Eric P. Xing Scribes: Fattaneh Jabbari, Eric Lei, Evan Shapiro 1 Introduction Probabilistic inference is one of
10-708: Probabilistic Graphical Models, Spring 2015 12 : Variational Inference I Lecturer: Eric P. Xing Scribes: Fattaneh Jabbari, Eric Lei, Evan Shapiro 1 Introduction Probabilistic inference is one of
Respecting Markov Equivalence in Computing Posterior Probabilities of Causal Graphical Features
 Proceedings of the Twenty-Fourth AAAI Conference on Artificial Intelligence (AAAI-10) Respecting Markov Equivalence in Computing Posterior Probabilities of Causal Graphical Features Eun Yong Kang Department
Proceedings of the Twenty-Fourth AAAI Conference on Artificial Intelligence (AAAI-10) Respecting Markov Equivalence in Computing Posterior Probabilities of Causal Graphical Features Eun Yong Kang Department
Weighting Expert Opinions in Group Decision Making for the Influential Effects between Variables in a Bayesian Network Model
 Weighting Expert Opinions in Group Decision Making for the Influential Effects between Variables in a Bayesian Network Model Nipat Jongsawat 1, Wichian Premchaiswadi 2 Graduate School of Information Technology
Weighting Expert Opinions in Group Decision Making for the Influential Effects between Variables in a Bayesian Network Model Nipat Jongsawat 1, Wichian Premchaiswadi 2 Graduate School of Information Technology
Probabilistic Graphical Models
 Probabilistic Graphical Models Lecture 5 Bayesian Learning of Bayesian Networks CS/CNS/EE 155 Andreas Krause Announcements Recitations: Every Tuesday 4-5:30 in 243 Annenberg Homework 1 out. Due in class
Probabilistic Graphical Models Lecture 5 Bayesian Learning of Bayesian Networks CS/CNS/EE 155 Andreas Krause Announcements Recitations: Every Tuesday 4-5:30 in 243 Annenberg Homework 1 out. Due in class
Bayesian Networks. Motivation
 Bayesian Networks Computer Sciences 760 Spring 2014 http://pages.cs.wisc.edu/~dpage/cs760/ Motivation Assume we have five Boolean variables,,,, The joint probability is,,,, How many state configurations
Bayesian Networks Computer Sciences 760 Spring 2014 http://pages.cs.wisc.edu/~dpage/cs760/ Motivation Assume we have five Boolean variables,,,, The joint probability is,,,, How many state configurations
Series 7, May 22, 2018 (EM Convergence)
 Exercises Introduction to Machine Learning SS 2018 Series 7, May 22, 2018 (EM Convergence) Institute for Machine Learning Dept. of Computer Science, ETH Zürich Prof. Dr. Andreas Krause Web: https://las.inf.ethz.ch/teaching/introml-s18
Exercises Introduction to Machine Learning SS 2018 Series 7, May 22, 2018 (EM Convergence) Institute for Machine Learning Dept. of Computer Science, ETH Zürich Prof. Dr. Andreas Krause Web: https://las.inf.ethz.ch/teaching/introml-s18
Learning Bayes Net Structures
 Learning Bayes Net Structures KF, Chapter 15 15.5 (RN, Chapter 20) Some material taken from C Guesterin (CMU), K Murphy (UBC) 1 2 Learning Bayes Nets Known Structure Unknown Data Complete Missing Easy
Learning Bayes Net Structures KF, Chapter 15 15.5 (RN, Chapter 20) Some material taken from C Guesterin (CMU), K Murphy (UBC) 1 2 Learning Bayes Nets Known Structure Unknown Data Complete Missing Easy
ECE521 Tutorial 11. Topic Review. ECE521 Winter Credits to Alireza Makhzani, Alex Schwing, Rich Zemel and TAs for slides. ECE521 Tutorial 11 / 4
 ECE52 Tutorial Topic Review ECE52 Winter 206 Credits to Alireza Makhzani, Alex Schwing, Rich Zemel and TAs for slides ECE52 Tutorial ECE52 Winter 206 Credits to Alireza / 4 Outline K-means, PCA 2 Bayesian
ECE52 Tutorial Topic Review ECE52 Winter 206 Credits to Alireza Makhzani, Alex Schwing, Rich Zemel and TAs for slides ECE52 Tutorial ECE52 Winter 206 Credits to Alireza / 4 Outline K-means, PCA 2 Bayesian
On the Complexity of Strong and Epistemic Credal Networks
 On the Complexity of Strong and Epistemic Credal Networks Denis D. Mauá Cassio P. de Campos Alessio Benavoli Alessandro Antonucci Istituto Dalle Molle di Studi sull Intelligenza Artificiale (IDSIA), Manno-Lugano,
On the Complexity of Strong and Epistemic Credal Networks Denis D. Mauá Cassio P. de Campos Alessio Benavoli Alessandro Antonucci Istituto Dalle Molle di Studi sull Intelligenza Artificiale (IDSIA), Manno-Lugano,
Unsupervised machine learning
 Chapter 9 Unsupervised machine learning Unsupervised machine learning (a.k.a. cluster analysis) is a set of methods to assign objects into clusters under a predefined distance measure when class labels
Chapter 9 Unsupervised machine learning Unsupervised machine learning (a.k.a. cluster analysis) is a set of methods to assign objects into clusters under a predefined distance measure when class labels
GLOBEX Bioinformatics (Summer 2015) Genetic networks and gene expression data
 GLOBEX Bioinformatics (Summer 2015) Genetic networks and gene expression data 1 Gene Networks Definition: A gene network is a set of molecular components, such as genes and proteins, and interactions between
GLOBEX Bioinformatics (Summer 2015) Genetic networks and gene expression data 1 Gene Networks Definition: A gene network is a set of molecular components, such as genes and proteins, and interactions between
Readings: K&F: 16.3, 16.4, Graphical Models Carlos Guestrin Carnegie Mellon University October 6 th, 2008
 Readings: K&F: 16.3, 16.4, 17.3 Bayesian Param. Learning Bayesian Structure Learning Graphical Models 10708 Carlos Guestrin Carnegie Mellon University October 6 th, 2008 10-708 Carlos Guestrin 2006-2008
Readings: K&F: 16.3, 16.4, 17.3 Bayesian Param. Learning Bayesian Structure Learning Graphical Models 10708 Carlos Guestrin Carnegie Mellon University October 6 th, 2008 10-708 Carlos Guestrin 2006-2008
An Empirical Investigation of the K2 Metric
 An Empirical Investigation of the K2 Metric Christian Borgelt and Rudolf Kruse Department of Knowledge Processing and Language Engineering Otto-von-Guericke-University of Magdeburg Universitätsplatz 2,
An Empirical Investigation of the K2 Metric Christian Borgelt and Rudolf Kruse Department of Knowledge Processing and Language Engineering Otto-von-Guericke-University of Magdeburg Universitätsplatz 2,
Embellishing a Bayesian Network using a Chain Event Graph
 Embellishing a Bayesian Network using a Chain Event Graph L. M. Barclay J. L. Hutton J. Q. Smith University of Warwick 4th Annual Conference of the Australasian Bayesian Network Modelling Society Example
Embellishing a Bayesian Network using a Chain Event Graph L. M. Barclay J. L. Hutton J. Q. Smith University of Warwick 4th Annual Conference of the Australasian Bayesian Network Modelling Society Example
Learning of Bayesian Network Structure from Massive Datasets: The Sparse Candidate Algorithm
 Learning of Bayesian Network Structure from Massive Datasets: The Sparse Candidate Algorithm Nir Friedman Institute of Computer Science Hebrew University Jerusalem, 91904, ISRAEL nir@cs.huji.ac.il Iftach
Learning of Bayesian Network Structure from Massive Datasets: The Sparse Candidate Algorithm Nir Friedman Institute of Computer Science Hebrew University Jerusalem, 91904, ISRAEL nir@cs.huji.ac.il Iftach
Lars Schmidt-Thieme, Information Systems and Machine Learning Lab (ISMLL), Institute BW/WI & Institute for Computer Science, University of Hildesheim
 Course on Bayesian Networks, summer term 2010 0/42 Bayesian Networks Bayesian Networks 11. Structure Learning / Constrained-based Lars Schmidt-Thieme Information Systems and Machine Learning Lab (ISMLL)
Course on Bayesian Networks, summer term 2010 0/42 Bayesian Networks Bayesian Networks 11. Structure Learning / Constrained-based Lars Schmidt-Thieme Information Systems and Machine Learning Lab (ISMLL)
Comparison of Shannon, Renyi and Tsallis Entropy used in Decision Trees
 Comparison of Shannon, Renyi and Tsallis Entropy used in Decision Trees Tomasz Maszczyk and W lodzis law Duch Department of Informatics, Nicolaus Copernicus University Grudzi adzka 5, 87-100 Toruń, Poland
Comparison of Shannon, Renyi and Tsallis Entropy used in Decision Trees Tomasz Maszczyk and W lodzis law Duch Department of Informatics, Nicolaus Copernicus University Grudzi adzka 5, 87-100 Toruń, Poland
Phylogenetic Networks, Trees, and Clusters
 Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Supplementary material to Structure Learning of Linear Gaussian Structural Equation Models with Weak Edges
 Supplementary material to Structure Learning of Linear Gaussian Structural Equation Models with Weak Edges 1 PRELIMINARIES Two vertices X i and X j are adjacent if there is an edge between them. A path
Supplementary material to Structure Learning of Linear Gaussian Structural Equation Models with Weak Edges 1 PRELIMINARIES Two vertices X i and X j are adjacent if there is an edge between them. A path
On Finding Optimal Polytrees
 Serge Gaspers The University of New South Wales and Vienna University of Technology gaspers@kr.tuwien.ac.at Mathieu Liedloff Université d Orléans mathieu.liedloff@univ-orleans.fr On Finding Optimal Polytrees
Serge Gaspers The University of New South Wales and Vienna University of Technology gaspers@kr.tuwien.ac.at Mathieu Liedloff Université d Orléans mathieu.liedloff@univ-orleans.fr On Finding Optimal Polytrees
Inference in Hybrid Bayesian Networks Using Mixtures of Gaussians
 428 SHENOY UAI 2006 Inference in Hybrid Bayesian Networks Using Mixtures of Gaussians Prakash P. Shenoy University of Kansas School of Business 1300 Sunnyside Ave, Summerfield Hall Lawrence, KS 66045-7585
428 SHENOY UAI 2006 Inference in Hybrid Bayesian Networks Using Mixtures of Gaussians Prakash P. Shenoy University of Kansas School of Business 1300 Sunnyside Ave, Summerfield Hall Lawrence, KS 66045-7585
Dynamic importance sampling in Bayesian networks using factorisation of probability trees
 Dynamic importance sampling in Bayesian networks using factorisation of probability trees Irene Martínez Department of Languages and Computation University of Almería Almería, 4, Spain Carmelo Rodríguez
Dynamic importance sampling in Bayesian networks using factorisation of probability trees Irene Martínez Department of Languages and Computation University of Almería Almería, 4, Spain Carmelo Rodríguez
A Branch-and-Bound Algorithm for MDL Learning Bayesian Networks
 580 UNCERTAINTY IN ARTIFICIAL INTELLIGENCE PROCEEDINGS 000 A Branch-and-Bound Algorithm for MDL Learning Bayesian Networks Jin Tian Cognitive Systems Laboratory Computer Science Department University of
580 UNCERTAINTY IN ARTIFICIAL INTELLIGENCE PROCEEDINGS 000 A Branch-and-Bound Algorithm for MDL Learning Bayesian Networks Jin Tian Cognitive Systems Laboratory Computer Science Department University of
Learning Bayesian networks
 1 Lecture topics: Learning Bayesian networks from data maximum likelihood, BIC Bayesian, marginal likelihood Learning Bayesian networks There are two problems we have to solve in order to estimate Bayesian
1 Lecture topics: Learning Bayesian networks from data maximum likelihood, BIC Bayesian, marginal likelihood Learning Bayesian networks There are two problems we have to solve in order to estimate Bayesian
The Necessity of Bounded Treewidth for Efficient Inference in Bayesian Networks
 The Necessity of Bounded Treewidth for Efficient Inference in Bayesian Networks Johan H.P. Kwisthout and Hans L. Bodlaender and L.C. van der Gaag 1 Abstract. Algorithms for probabilistic inference in Bayesian
The Necessity of Bounded Treewidth for Efficient Inference in Bayesian Networks Johan H.P. Kwisthout and Hans L. Bodlaender and L.C. van der Gaag 1 Abstract. Algorithms for probabilistic inference in Bayesian
Probabilistic Graphical Models
 Parameter Estimation December 14, 2015 Overview 1 Motivation 2 3 4 What did we have so far? 1 Representations: how do we model the problem? (directed/undirected). 2 Inference: given a model and partially
Parameter Estimation December 14, 2015 Overview 1 Motivation 2 3 4 What did we have so far? 1 Representations: how do we model the problem? (directed/undirected). 2 Inference: given a model and partially
A Memetic Approach to Bayesian Network Structure Learning
 A Memetic Approach to Bayesian Network Structure Learning Author 1 1, Author 2 2, Author 3 3, and Author 4 4 1 Institute 1 author1@institute1.com 2 Institute 2 author2@institute2.com 3 Institute 3 author3@institute3.com
A Memetic Approach to Bayesian Network Structure Learning Author 1 1, Author 2 2, Author 3 3, and Author 4 4 1 Institute 1 author1@institute1.com 2 Institute 2 author2@institute2.com 3 Institute 3 author3@institute3.com
Lecture 9: Bayesian Learning
 Lecture 9: Bayesian Learning Cognitive Systems II - Machine Learning Part II: Special Aspects of Concept Learning Bayes Theorem, MAL / ML hypotheses, Brute-force MAP LEARNING, MDL principle, Bayes Optimal
Lecture 9: Bayesian Learning Cognitive Systems II - Machine Learning Part II: Special Aspects of Concept Learning Bayes Theorem, MAL / ML hypotheses, Brute-force MAP LEARNING, MDL principle, Bayes Optimal
Mixtures of Gaussians with Sparse Structure
 Mixtures of Gaussians with Sparse Structure Costas Boulis 1 Abstract When fitting a mixture of Gaussians to training data there are usually two choices for the type of Gaussians used. Either diagonal or
Mixtures of Gaussians with Sparse Structure Costas Boulis 1 Abstract When fitting a mixture of Gaussians to training data there are usually two choices for the type of Gaussians used. Either diagonal or
The Parameterized Complexity of Approximate Inference in Bayesian Networks
 JMLR: Workshop and Conference Proceedings vol 52, 264-274, 2016 PGM 2016 The Parameterized Complexity of Approximate Inference in Bayesian Networks Johan Kwisthout Radboud University Donders Institute
JMLR: Workshop and Conference Proceedings vol 52, 264-274, 2016 PGM 2016 The Parameterized Complexity of Approximate Inference in Bayesian Networks Johan Kwisthout Radboud University Donders Institute
Motivation. Bayesian Networks in Epistemology and Philosophy of Science Lecture. Overview. Organizational Issues
 Bayesian Networks in Epistemology and Philosophy of Science Lecture 1: Bayesian Networks Center for Logic and Philosophy of Science Tilburg University, The Netherlands Formal Epistemology Course Northern
Bayesian Networks in Epistemology and Philosophy of Science Lecture 1: Bayesian Networks Center for Logic and Philosophy of Science Tilburg University, The Netherlands Formal Epistemology Course Northern
Chapter 3 Deterministic planning
 Chapter 3 Deterministic planning In this chapter we describe a number of algorithms for solving the historically most important and most basic type of planning problem. Two rather strong simplifying assumptions
Chapter 3 Deterministic planning In this chapter we describe a number of algorithms for solving the historically most important and most basic type of planning problem. Two rather strong simplifying assumptions
Being Bayesian About Network Structure
 Being Bayesian About Network Structure A Bayesian Approach to Structure Discovery in Bayesian Networks Nir Friedman (nir@cs.huji.ac.il) School of Computer Science & Engineering Hebrew University Jerusalem,
Being Bayesian About Network Structure A Bayesian Approach to Structure Discovery in Bayesian Networks Nir Friedman (nir@cs.huji.ac.il) School of Computer Science & Engineering Hebrew University Jerusalem,
The Bayesian Structural EM Algorithm
 The Bayesian Structural EM Algorithm Nir Friedman Computer Science Division, 387 Soda Hall University of California, Berkeley, CA 9470 nir@cs.berkeley.edu Abstract In recent years there has been a flurry
The Bayesian Structural EM Algorithm Nir Friedman Computer Science Division, 387 Soda Hall University of California, Berkeley, CA 9470 nir@cs.berkeley.edu Abstract In recent years there has been a flurry
PBIL-RS: Effective Repeated Search in PBIL-based Bayesian Network Learning
 International Journal of Informatics Society, VOL.8, NO.1 (2016) 15-23 15 PBIL-RS: Effective Repeated Search in PBIL-based Bayesian Network Learning Yuma Yamanaka, Takatoshi Fujiki, Sho Fukuda, and Takuya
International Journal of Informatics Society, VOL.8, NO.1 (2016) 15-23 15 PBIL-RS: Effective Repeated Search in PBIL-based Bayesian Network Learning Yuma Yamanaka, Takatoshi Fujiki, Sho Fukuda, and Takuya
An Introduction to Bayesian Machine Learning
 1 An Introduction to Bayesian Machine Learning José Miguel Hernández-Lobato Department of Engineering, Cambridge University April 8, 2013 2 What is Machine Learning? The design of computational systems
1 An Introduction to Bayesian Machine Learning José Miguel Hernández-Lobato Department of Engineering, Cambridge University April 8, 2013 2 What is Machine Learning? The design of computational systems
Maximum likelihood bounded tree-width Markov networks
 Artificial Intelligence 143 (2003) 123 138 www.elsevier.com/locate/artint Maximum likelihood bounded tree-width Markov networks Nathan Srebro Department of Electrical Engineering and Computer Science,
Artificial Intelligence 143 (2003) 123 138 www.elsevier.com/locate/artint Maximum likelihood bounded tree-width Markov networks Nathan Srebro Department of Electrical Engineering and Computer Science,
Undirected Graphical Models
 Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Properties Properties 3 Generative vs. Conditional
Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Properties Properties 3 Generative vs. Conditional
Chris Bishop s PRML Ch. 8: Graphical Models
 Chris Bishop s PRML Ch. 8: Graphical Models January 24, 2008 Introduction Visualize the structure of a probabilistic model Design and motivate new models Insights into the model s properties, in particular
Chris Bishop s PRML Ch. 8: Graphical Models January 24, 2008 Introduction Visualize the structure of a probabilistic model Design and motivate new models Insights into the model s properties, in particular
Graphical Models and Independence Models
 Graphical Models and Independence Models Yunshu Liu ASPITRG Research Group 2014-03-04 References: [1]. Steffen Lauritzen, Graphical Models, Oxford University Press, 1996 [2]. Christopher M. Bishop, Pattern
Graphical Models and Independence Models Yunshu Liu ASPITRG Research Group 2014-03-04 References: [1]. Steffen Lauritzen, Graphical Models, Oxford University Press, 1996 [2]. Christopher M. Bishop, Pattern
CS Lecture 3. More Bayesian Networks
 CS 6347 Lecture 3 More Bayesian Networks Recap Last time: Complexity challenges Representing distributions Computing probabilities/doing inference Introduction to Bayesian networks Today: D-separation,
CS 6347 Lecture 3 More Bayesian Networks Recap Last time: Complexity challenges Representing distributions Computing probabilities/doing inference Introduction to Bayesian networks Today: D-separation,
The size of decision table can be understood in terms of both cardinality of A, denoted by card (A), and the number of equivalence classes of IND (A),
 Attribute Set Decomposition of Decision Tables Dominik Slezak Warsaw University Banacha 2, 02-097 Warsaw Phone: +48 (22) 658-34-49 Fax: +48 (22) 658-34-48 Email: slezak@alfa.mimuw.edu.pl ABSTRACT: Approach
Attribute Set Decomposition of Decision Tables Dominik Slezak Warsaw University Banacha 2, 02-097 Warsaw Phone: +48 (22) 658-34-49 Fax: +48 (22) 658-34-48 Email: slezak@alfa.mimuw.edu.pl ABSTRACT: Approach
15-780: Graduate Artificial Intelligence. Bayesian networks: Construction and inference
 15-780: Graduate Artificial Intelligence ayesian networks: Construction and inference ayesian networks: Notations ayesian networks are directed acyclic graphs. Conditional probability tables (CPTs) P(Lo)
15-780: Graduate Artificial Intelligence ayesian networks: Construction and inference ayesian networks: Notations ayesian networks are directed acyclic graphs. Conditional probability tables (CPTs) P(Lo)
A Result of Vapnik with Applications
 A Result of Vapnik with Applications Martin Anthony Department of Statistical and Mathematical Sciences London School of Economics Houghton Street London WC2A 2AE, U.K. John Shawe-Taylor Department of
A Result of Vapnik with Applications Martin Anthony Department of Statistical and Mathematical Sciences London School of Economics Houghton Street London WC2A 2AE, U.K. John Shawe-Taylor Department of
When Discriminative Learning of Bayesian Network Parameters Is Easy
 Pp. 491 496 in Proceedings of the Eighteenth International Joint Conference on Artificial Intelligence (IJCAI 2003), edited by G. Gottlob and T. Walsh. Morgan Kaufmann, 2003. When Discriminative Learning
Pp. 491 496 in Proceedings of the Eighteenth International Joint Conference on Artificial Intelligence (IJCAI 2003), edited by G. Gottlob and T. Walsh. Morgan Kaufmann, 2003. When Discriminative Learning
13 : Variational Inference: Loopy Belief Propagation and Mean Field
 10-708: Probabilistic Graphical Models 10-708, Spring 2012 13 : Variational Inference: Loopy Belief Propagation and Mean Field Lecturer: Eric P. Xing Scribes: Peter Schulam and William Wang 1 Introduction
10-708: Probabilistic Graphical Models 10-708, Spring 2012 13 : Variational Inference: Loopy Belief Propagation and Mean Field Lecturer: Eric P. Xing Scribes: Peter Schulam and William Wang 1 Introduction
Efficient Sensitivity Analysis in Hidden Markov Models
 Efficient Sensitivity Analysis in Hidden Markov Models Silja Renooij Department of Information and Computing Sciences, Utrecht University P.O. Box 80.089, 3508 TB Utrecht, The Netherlands silja@cs.uu.nl
Efficient Sensitivity Analysis in Hidden Markov Models Silja Renooij Department of Information and Computing Sciences, Utrecht University P.O. Box 80.089, 3508 TB Utrecht, The Netherlands silja@cs.uu.nl
Bayesian Networks. B. Inference / B.3 Approximate Inference / Sampling
 Bayesian etworks B. Inference / B.3 Approximate Inference / Sampling Lars Schmidt-Thieme Information Systems and Machine Learning Lab (ISMLL) Institute of Computer Science University of Hildesheim http://www.ismll.uni-hildesheim.de
Bayesian etworks B. Inference / B.3 Approximate Inference / Sampling Lars Schmidt-Thieme Information Systems and Machine Learning Lab (ISMLL) Institute of Computer Science University of Hildesheim http://www.ismll.uni-hildesheim.de
