I wonder about Trees Robert Frost
|
|
|
- Gabriel Blake
- 5 years ago
- Views:
Transcription
1 I wonder about Trees Robert Frost We wonder about Robert Frost - Trees Hey! How come no turtles in this tree?
2 Phylogenetic Trees From Darwin s notebooks, 1837
3
4 Really?
5 The only figure in On the Origin of Species by Natural Selection (Darwin, 1859) From the 6 th edition (1872): The affinities of all the beings of the same class have sometimes been represented by a great tree. I believe this simile largely speaks the truth. The green and budding twigs may represent existing species; and those produced during former years may represent the long succession of extinct species.
6 More recursion on trees: mrca West Dorm Groody ( W ) >>> find( L, Groodies) True X Y Z East Dorm Groody ( E ) Linde Dorm Groody ( L ) Case Groody ( C ) >>> mrca("l", "E", Groodies) # use find as a helper! 'Z' >>> mrca("w", "E", Groodies) 'Y' >>> mrca("w", "C", Groodies) 'X' >>> mrca("w", "Prof. Wu", Groodies) None
7 mrca X Y Z West Dorm Groody ( W ) East Dorm Groody ( E ) Linde Dorm Groody ( L ) def mrca(species1,species2,tree): """Return the name of the most recent commmon ancestor of species1 and species2. If there isn't one, return None.""" Case Groody ( C ) Notes
8 mrca X Y Z West Dorm Groody ( W ) East Dorm Groody ( E ) Linde Dorm Groody ( L ) def mrca(species1,species2,tree): """Return the name of the most recent commmon ancestor of species1 and species2. If there isn't one, return None.""" if Tree[1] == (): return None elif not find(species1,tree) or not find(species2,tree): return None else: if find(species1, Tree[1]) and find(species2, Tree[1]): return mrca(species1,species2, Tree[1]) elif find(species1, Tree[2]) and find(species2, Tree[2]): return mrca(species1,species2, Tree[2]) else: return Tree[0] Case Groody ( C )
9 Phylogenetic Reconstruction Input: DNA or protein sequences for each species Output: Species tree D. Desjardin, K. Peay, T. Bruns, Mycologia 103(5), 2011
10
11 Distance-Based Approach Groody CATCAACCAGTGACCAGTATAGGACGCCC Froody CAACACTCAGTGACAAGTCTAGCACGCCC Snoody AATCGCCCGGCGTCAGGCATAGCTAGCGC G F S G F S
12 Distance-Based Approach Current time ( time 0 ) Snoody G F S G F S Groody Froody (6.0, ('Snoody', (), ()), (3.0, ('Groody', (), ()), ('Froody', (), ()) ) )
13 Unweighted Pair Group Method with Arithmetic Mean (UPGMA) Algorithm Groody Froody Snoody G F S G F S
14 Try This One Aoody Boody Coody Doody Eoody A B C D E A 0 B 4 0 C D E This algorithm s bark is worse than it s bite. The matrix is symmetric, so we just need to keep the bottom or top half! Worksheet
15
16
17 DEMO! I wood love to see this in action!
18 Implementing UPGMA 1 Groody 2 Froody 3 Snoody Let s see this in the provided mitodata.py file N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) groodieslist = [N1, N2, N3] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0}
19 N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) groodieslist = [N1, N2, N3] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0} 1. For all pairs of trees in the groodieslist, find the closest pair
20 N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) groodieslist = [N1, N2, N3] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0} 1. For all pairs of trees in the groodieslist, find the closest pair N1 = ("Groody", (), ()) N2 = ("Froody", (), ())
21 N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) groodieslist = [N1, N2, N3] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0} 1. For all pairs of trees in the groodieslist, find the closest pair N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) 2. Remove those from the groodieslist groodylist.remove(n1) groodylist.remove(n2)
22 N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) groodieslist = [N1, N2, N3] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0} 1. For all pairs of trees in the groodieslist, find the closest pair N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) 2. Remove those from the groodieslist 3. Make a new tree by joining these two trees
23 N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) groodieslist = [N1, N2, N3] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0} 1. For all pairs of trees in the groodieslist, find the closest pair N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) 2. Remove those from the groodieslist 3. Make a new tree by joining these two trees (3.0, N1, N2) = (3.0, ("Groody", (), ()), ("Froody", (), ()))
24 N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) N1 N2 groodieslist = [N3, (3.0, ("Groody", (), ()), ("Froody", (), ()))] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0} 1. For all pairs of trees in the groodieslist, find the closest pair N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) 2. Remove those from the groodieslist 3. Make a new tree by joining these two trees (3.0, N1, N2) = (3.0, ("Groody", (), ()), ("Froody", (), ())) 4. Add this new tree to the groodieslist
25 N1 = ("Groody", (), ()) N2 = ("Froody", (), ()) N3 = ("Snoody", (), ()) N1 N2 groodieslist = [N3, (3.0, ("Groody", (), ()), ("Froody", (), ()))] groodiesmatrix = {(N1, N1):0, (N1, N2):6, (N1, N3):12, (N2, N1):6, (N2, N2):0, (N2, N3):12, (N3,N1):12, (N3,N2):12, (N3,N3):0} 5. Update the distance matrix We are still holding on to N1 and N2, even though we have removed them from the groodieslist!
26 Implementing UPGMA findclosestpair( specieslist, Distances ): Takes a list of species trees and the distance dictionary as input and returns a tuple (X, Y) where X and Y are in the list and have the minimum distance between any two items in the list. updatedist( specieslist, Distances, newtree): Takes a list of species trees, the distance dictionary, and a newtree that was just formed by merging two trees found by findclosestpair. Those two trees can be found by looking inside newtree. Those two trees are removed from the distance dictionary and the newtree is added to the dictionary. upgma( specieslist, Distances): returns the phylogenetic tree constructed by the UPGMA algorithm findclosestpair: 7 lines updatedist: 8 lines upgma: 12 lines
27 Updating the distance dictionary: a subtlety a b c d a 0 b 4 0 c d a,b c d a,b 0 c 6 0 d a,b,c d a,b,c 0 d??? c a b
28 Updating the distance dictionary: a subtlety a b c d a 0 b 4 0 c d a,b c d a,b 0 c 6 0 d a,b,c d a,b,c 0 d 11 0 When we combine distances between nodes with different numbers of leaves, we should weight by the number of leaves. 2 leaves 1 leaf = 3 total a,b c 2/3 * /3 * 9 = c a b
29 Updating the distance dictionary: a subtlety a b c d a 0 b 4 0 c d a,b c d a,b 0 c 6 0 d a,b,c d a,b,c 0 d 11 0 When we combine distances between nodes with different numbers of leaves, we should weight by the number of leaves. 2 leaves 1 leaf = 3 total a,b c 2/3 * /3 * 9 = c a b newtree = ( Anc, T1, T2) Distance(newTree, T3) = Distance(T1, T3) x leafcount (T1)/leafCount(newTree) + Distance(T2, T3) x leafcount(t2)/leafcount(newtree)
30 Inferring time 3 A B C D The scale function you write in lab will be useful
31 Rhea Ostrich
32 UPGMA assumes a molecular clock G F S G F S
33 A note on biogeography/migrations
Trees and Human Evolution
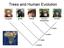 Trees and Human Evolution orangutan gorilla human common chimpanzee bonobo ~3 MYA ~6 MYA ~7 MYA ~15 MYA Learning Goals Introduce biological question Describe how data is stored in memory (just a peek)
Trees and Human Evolution orangutan gorilla human common chimpanzee bonobo ~3 MYA ~6 MYA ~7 MYA ~15 MYA Learning Goals Introduce biological question Describe how data is stored in memory (just a peek)
C.DARWIN ( )
 C.DARWIN (1809-1882) LAMARCK Each evolutionary lineage has evolved, transforming itself, from a ancestor appeared by spontaneous generation DARWIN All organisms are historically interconnected. Their relationships
C.DARWIN (1809-1882) LAMARCK Each evolutionary lineage has evolved, transforming itself, from a ancestor appeared by spontaneous generation DARWIN All organisms are historically interconnected. Their relationships
Phylogenetic trees 07/10/13
 Phylogenetic trees 07/10/13 A tree is the only figure to occur in On the Origin of Species by Charles Darwin. It is a graphical representation of the evolutionary relationships among entities that share
Phylogenetic trees 07/10/13 A tree is the only figure to occur in On the Origin of Species by Charles Darwin. It is a graphical representation of the evolutionary relationships among entities that share
Evolutionary Tree Analysis. Overview
 CSI/BINF 5330 Evolutionary Tree Analysis Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Distance-Based Evolutionary Tree Reconstruction Character-Based
CSI/BINF 5330 Evolutionary Tree Analysis Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Distance-Based Evolutionary Tree Reconstruction Character-Based
Algorithms in Bioinformatics
 Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Distance Methods Character Methods
Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Distance Methods Character Methods
Theory of Evolution Charles Darwin
 Theory of Evolution Charles arwin 858-59: Origin of Species 5 year voyage of H.M.S. eagle (83-36) Populations have variations. Natural Selection & Survival of the fittest: nature selects best adapted varieties
Theory of Evolution Charles arwin 858-59: Origin of Species 5 year voyage of H.M.S. eagle (83-36) Populations have variations. Natural Selection & Survival of the fittest: nature selects best adapted varieties
Inferring Phylogenetic Trees. Distance Approaches. Representing distances. in rooted and unrooted trees. The distance approach to phylogenies
 Inferring Phylogenetic Trees Distance Approaches Representing distances in rooted and unrooted trees The distance approach to phylogenies given: an n n matrix M where M ij is the distance between taxa
Inferring Phylogenetic Trees Distance Approaches Representing distances in rooted and unrooted trees The distance approach to phylogenies given: an n n matrix M where M ij is the distance between taxa
EVOLUTIONARY DISTANCES
 EVOLUTIONARY DISTANCES FROM STRINGS TO TREES Luca Bortolussi 1 1 Dipartimento di Matematica ed Informatica Università degli studi di Trieste luca@dmi.units.it Trieste, 14 th November 2007 OUTLINE 1 STRINGS:
EVOLUTIONARY DISTANCES FROM STRINGS TO TREES Luca Bortolussi 1 1 Dipartimento di Matematica ed Informatica Università degli studi di Trieste luca@dmi.units.it Trieste, 14 th November 2007 OUTLINE 1 STRINGS:
BINF6201/8201. Molecular phylogenetic methods
 BINF60/80 Molecular phylogenetic methods 0-7-06 Phylogenetics Ø According to the evolutionary theory, all life forms on this planet are related to one another by descent. Ø Traditionally, phylogenetics
BINF60/80 Molecular phylogenetic methods 0-7-06 Phylogenetics Ø According to the evolutionary theory, all life forms on this planet are related to one another by descent. Ø Traditionally, phylogenetics
Phylogenetic Analysis. Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center
 Phylogenetic Analysis Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center Outline Basic Concepts Tree Construction Methods Distance-based methods
Phylogenetic Analysis Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center Outline Basic Concepts Tree Construction Methods Distance-based methods
Phylogenetic Trees. Phylogenetic Trees Five. Phylogeny: Inference Tool. Phylogeny Terminology. Picture of Last Quagga. Importance of Phylogeny 5.
 Five Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu v Distance Methods v Character Methods v Molecular Clock v UPGMA v Maximum Parsimony
Five Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu v Distance Methods v Character Methods v Molecular Clock v UPGMA v Maximum Parsimony
Multiple Sequence Alignment. Sequences
 Multiple Sequence Alignment Sequences > YOR020c mstllksaksivplmdrvlvqrikaqaktasglylpe knveklnqaevvavgpgftdangnkvvpqvkvgdqvl ipqfggstiklgnddevilfrdaeilakiakd > crassa mattvrsvksliplldrvlvqrvkaeaktasgiflpe
Multiple Sequence Alignment Sequences > YOR020c mstllksaksivplmdrvlvqrikaqaktasglylpe knveklnqaevvavgpgftdangnkvvpqvkvgdqvl ipqfggstiklgnddevilfrdaeilakiakd > crassa mattvrsvksliplldrvlvqrvkaeaktasgiflpe
Evolutionary Trees. Evolutionary tree. To describe the evolutionary relationship among species A 3 A 2 A 4. R.C.T. Lee and Chin Lung Lu
 Evolutionary Trees R.C.T. Lee and Chin Lung Lu CS 5313 Algorithms for Molecular Biology Evolutionary Trees p.1 Evolutionary tree To describe the evolutionary relationship among species Root A 3 Bifurcating
Evolutionary Trees R.C.T. Lee and Chin Lung Lu CS 5313 Algorithms for Molecular Biology Evolutionary Trees p.1 Evolutionary tree To describe the evolutionary relationship among species Root A 3 Bifurcating
Phylogenetic Tree Reconstruction
 I519 Introduction to Bioinformatics, 2011 Phylogenetic Tree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Evolution theory Speciation Evolution of new organisms is driven
I519 Introduction to Bioinformatics, 2011 Phylogenetic Tree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Evolution theory Speciation Evolution of new organisms is driven
"Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky
 MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
A (short) introduction to phylogenetics
 A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
Tree Building Activity
 Tree Building Activity Introduction In this activity, you will construct phylogenetic trees using a phenotypic similarity (cartoon microbe pictures) and genotypic similarity (real microbe sequences). For
Tree Building Activity Introduction In this activity, you will construct phylogenetic trees using a phenotypic similarity (cartoon microbe pictures) and genotypic similarity (real microbe sequences). For
Molecular Evolution and Phylogenetic Tree Reconstruction
 1 4 Molecular Evolution and Phylogenetic Tree Reconstruction 3 2 5 1 4 2 3 5 Orthology, Paralogy, Inparalogs, Outparalogs Phylogenetic Trees Nodes: species Edges: time of independent evolution Edge length
1 4 Molecular Evolution and Phylogenetic Tree Reconstruction 3 2 5 1 4 2 3 5 Orthology, Paralogy, Inparalogs, Outparalogs Phylogenetic Trees Nodes: species Edges: time of independent evolution Edge length
Constructing Evolutionary/Phylogenetic Trees
 Constructing Evolutionary/Phylogenetic Trees 2 broad categories: istance-based methods Ultrametric Additive: UPGMA Transformed istance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
Constructing Evolutionary/Phylogenetic Trees 2 broad categories: istance-based methods Ultrametric Additive: UPGMA Transformed istance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
What is Phylogenetics
 What is Phylogenetics Phylogenetics is the area of research concerned with finding the genetic connections and relationships between species. The basic idea is to compare specific characters (features)
What is Phylogenetics Phylogenetics is the area of research concerned with finding the genetic connections and relationships between species. The basic idea is to compare specific characters (features)
CS5238 Combinatorial methods in bioinformatics 2003/2004 Semester 1. Lecture 8: Phylogenetic Tree Reconstruction: Distance Based - October 10, 2003
 CS5238 Combinatorial methods in bioinformatics 2003/2004 Semester 1 Lecture 8: Phylogenetic Tree Reconstruction: Distance Based - October 10, 2003 Lecturer: Wing-Kin Sung Scribe: Ning K., Shan T., Xiang
CS5238 Combinatorial methods in bioinformatics 2003/2004 Semester 1 Lecture 8: Phylogenetic Tree Reconstruction: Distance Based - October 10, 2003 Lecturer: Wing-Kin Sung Scribe: Ning K., Shan T., Xiang
Evidence of Evolution
 16.4 Evidence for Evolution Biogeography Biogeography - study of where organisms live, where they and ancestors lived. Two significant patterns: - closely related species separate in different climates.
16.4 Evidence for Evolution Biogeography Biogeography - study of where organisms live, where they and ancestors lived. Two significant patterns: - closely related species separate in different climates.
THEORY. Based on sequence Length According to the length of sequence being compared it is of following two types
 Exp 11- THEORY Sequence Alignment is a process of aligning two sequences to achieve maximum levels of identity between them. This help to derive functional, structural and evolutionary relationships between
Exp 11- THEORY Sequence Alignment is a process of aligning two sequences to achieve maximum levels of identity between them. This help to derive functional, structural and evolutionary relationships between
Plan: Evolutionary trees, characters. Perfect phylogeny Methods: NJ, parsimony, max likelihood, Quartet method
 Phylogeny 1 Plan: Phylogeny is an important subject. We have 2.5 hours. So I will teach all the concepts via one example of a chain letter evolution. The concepts we will discuss include: Evolutionary
Phylogeny 1 Plan: Phylogeny is an important subject. We have 2.5 hours. So I will teach all the concepts via one example of a chain letter evolution. The concepts we will discuss include: Evolutionary
COMP 355 Advanced Algorithms
 COMP 355 Advanced Algorithms Algorithm Design Review: Mathematical Background 1 Polynomial Running Time Brute force. For many non-trivial problems, there is a natural brute force search algorithm that
COMP 355 Advanced Algorithms Algorithm Design Review: Mathematical Background 1 Polynomial Running Time Brute force. For many non-trivial problems, there is a natural brute force search algorithm that
Page 1. Evolutionary Trees. Why build evolutionary tree? Outline
 Page Evolutionary Trees Russ. ltman MI S 7 Outline. Why build evolutionary trees?. istance-based vs. character-based methods. istance-based: Ultrametric Trees dditive Trees. haracter-based: Perfect phylogeny
Page Evolutionary Trees Russ. ltman MI S 7 Outline. Why build evolutionary trees?. istance-based vs. character-based methods. istance-based: Ultrametric Trees dditive Trees. haracter-based: Perfect phylogeny
Using Bioinformatics to Study Evolutionary Relationships Instructions
 3 Using Bioinformatics to Study Evolutionary Relationships Instructions Student Researcher Background: Making and Using Multiple Sequence Alignments One of the primary tasks of genetic researchers is comparing
3 Using Bioinformatics to Study Evolutionary Relationships Instructions Student Researcher Background: Making and Using Multiple Sequence Alignments One of the primary tasks of genetic researchers is comparing
COMP 355 Advanced Algorithms Algorithm Design Review: Mathematical Background
 COMP 355 Advanced Algorithms Algorithm Design Review: Mathematical Background 1 Polynomial Time Brute force. For many non-trivial problems, there is a natural brute force search algorithm that checks every
COMP 355 Advanced Algorithms Algorithm Design Review: Mathematical Background 1 Polynomial Time Brute force. For many non-trivial problems, there is a natural brute force search algorithm that checks every
Phylogenetics: Distance Methods. COMP Spring 2015 Luay Nakhleh, Rice University
 Phylogenetics: Distance Methods COMP 571 - Spring 2015 Luay Nakhleh, Rice University Outline Evolutionary models and distance corrections Distance-based methods Evolutionary Models and Distance Correction
Phylogenetics: Distance Methods COMP 571 - Spring 2015 Luay Nakhleh, Rice University Outline Evolutionary models and distance corrections Distance-based methods Evolutionary Models and Distance Correction
Phylogenetic inference
 Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Gel Electrophoresis. 10/28/0310/21/2003 CAP/CGS 5991 Lecture 10Lecture 9 1
 Gel Electrophoresis Used to measure the lengths of DNA fragments. When voltage is applied to DNA, different size fragments migrate to different distances (smaller ones travel farther). 10/28/0310/21/2003
Gel Electrophoresis Used to measure the lengths of DNA fragments. When voltage is applied to DNA, different size fragments migrate to different distances (smaller ones travel farther). 10/28/0310/21/2003
C3020 Molecular Evolution. Exercises #3: Phylogenetics
 C3020 Molecular Evolution Exercises #3: Phylogenetics Consider the following sequences for five taxa 1-5 and the known outgroup O, which has the ancestral states (note that sequence 3 has changed from
C3020 Molecular Evolution Exercises #3: Phylogenetics Consider the following sequences for five taxa 1-5 and the known outgroup O, which has the ancestral states (note that sequence 3 has changed from
Phylogenetic Trees. What They Are Why We Do It & How To Do It. Presented by Amy Harris Dr Brad Morantz
 Phylogenetic Trees What They Are Why We Do It & How To Do It Presented by Amy Harris Dr Brad Morantz Overview What is a phylogenetic tree Why do we do it How do we do it Methods and programs Parallels
Phylogenetic Trees What They Are Why We Do It & How To Do It Presented by Amy Harris Dr Brad Morantz Overview What is a phylogenetic tree Why do we do it How do we do it Methods and programs Parallels
Evolutionary trees. Describe the relationship between objects, e.g. species or genes
 Evolutionary trees Bonobo Chimpanzee Human Neanderthal Gorilla Orangutan Describe the relationship between objects, e.g. species or genes Early evolutionary studies The evolutionary relationships between
Evolutionary trees Bonobo Chimpanzee Human Neanderthal Gorilla Orangutan Describe the relationship between objects, e.g. species or genes Early evolutionary studies The evolutionary relationships between
Data Structures and Algorithms
 Data Structures and Algorithms Spring 2017-2018 Outline 1 Sorting Algorithms (contd.) Outline Sorting Algorithms (contd.) 1 Sorting Algorithms (contd.) Analysis of Quicksort Time to sort array of length
Data Structures and Algorithms Spring 2017-2018 Outline 1 Sorting Algorithms (contd.) Outline Sorting Algorithms (contd.) 1 Sorting Algorithms (contd.) Analysis of Quicksort Time to sort array of length
Theory of Evolution. Charles Darwin
 Theory of Evolution harles arwin 858-59: Origin of Species 5 year voyage of H.M.S. eagle (8-6) Populations have variations. Natural Selection & Survival of the fittest: nature selects best adapted varieties
Theory of Evolution harles arwin 858-59: Origin of Species 5 year voyage of H.M.S. eagle (8-6) Populations have variations. Natural Selection & Survival of the fittest: nature selects best adapted varieties
RECOVERING NORMAL NETWORKS FROM SHORTEST INTER-TAXA DISTANCE INFORMATION
 RECOVERING NORMAL NETWORKS FROM SHORTEST INTER-TAXA DISTANCE INFORMATION MAGNUS BORDEWICH, KATHARINA T. HUBER, VINCENT MOULTON, AND CHARLES SEMPLE Abstract. Phylogenetic networks are a type of leaf-labelled,
RECOVERING NORMAL NETWORKS FROM SHORTEST INTER-TAXA DISTANCE INFORMATION MAGNUS BORDEWICH, KATHARINA T. HUBER, VINCENT MOULTON, AND CHARLES SEMPLE Abstract. Phylogenetic networks are a type of leaf-labelled,
Phylogenetic inference: from sequences to trees
 W ESTFÄLISCHE W ESTFÄLISCHE W ILHELMS -U NIVERSITÄT NIVERSITÄT WILHELMS-U ÜNSTER MM ÜNSTER VOLUTIONARY FUNCTIONAL UNCTIONAL GENOMICS ENOMICS EVOLUTIONARY Bioinformatics 1 Phylogenetic inference: from sequences
W ESTFÄLISCHE W ESTFÄLISCHE W ILHELMS -U NIVERSITÄT NIVERSITÄT WILHELMS-U ÜNSTER MM ÜNSTER VOLUTIONARY FUNCTIONAL UNCTIONAL GENOMICS ENOMICS EVOLUTIONARY Bioinformatics 1 Phylogenetic inference: from sequences
Phylogeny: building the tree of life
 Phylogeny: building the tree of life Dr. Fayyaz ul Amir Afsar Minhas Department of Computer and Information Sciences Pakistan Institute of Engineering & Applied Sciences PO Nilore, Islamabad, Pakistan
Phylogeny: building the tree of life Dr. Fayyaz ul Amir Afsar Minhas Department of Computer and Information Sciences Pakistan Institute of Engineering & Applied Sciences PO Nilore, Islamabad, Pakistan
4/25/2009. Xi (Cici) Chen Texas Tech University. Ancient: folk taxonomy impulse to classify organisms Carl Linnaenus: binominal nomenclature (1735)
 Xi (Cici) Chen Texas Tech University Ancient: folk taxonomy impulse to classify y p y organisms Carl Linnaenus: binominal nomenclature (1735) 1 Darwin, On the origin of species The affinities of all the
Xi (Cici) Chen Texas Tech University Ancient: folk taxonomy impulse to classify y p y organisms Carl Linnaenus: binominal nomenclature (1735) 1 Darwin, On the origin of species The affinities of all the
CSCI1950 Z Computa4onal Methods for Biology Lecture 5
 CSCI1950 Z Computa4onal Methods for Biology Lecture 5 Ben Raphael February 6, 2009 hip://cs.brown.edu/courses/csci1950 z/ Alignment vs. Distance Matrix Mouse: ACAGTGACGCCACACACGT Gorilla: CCTGCGACGTAACAAACGC
CSCI1950 Z Computa4onal Methods for Biology Lecture 5 Ben Raphael February 6, 2009 hip://cs.brown.edu/courses/csci1950 z/ Alignment vs. Distance Matrix Mouse: ACAGTGACGCCACACACGT Gorilla: CCTGCGACGTAACAAACGC
6.001 SICP September? Procedural abstraction example: sqrt (define try (lambda (guess x) (if (good-enuf? guess x) guess (try (improve guess x) x))))
 September? 600-Introduction Trevor Darrell trevor@csail.mit.edu -D5 6.00 web page: http://sicp.csail.mit.edu/ section web page: http://www.csail.mit.edu/~trevor/600/ Abstractions Pairs and lists Common
September? 600-Introduction Trevor Darrell trevor@csail.mit.edu -D5 6.00 web page: http://sicp.csail.mit.edu/ section web page: http://www.csail.mit.edu/~trevor/600/ Abstractions Pairs and lists Common
Inferring phylogeny. Today s topics. Milestones of molecular evolution studies Contributions to molecular evolution
 Today s topics Inferring phylogeny Introduction! Distance methods! Parsimony method!"#$%&'(!)* +,-.'/01!23454(6!7!2845*0&4'9#6!:&454(6 ;?@AB=C?DEF Overview of phylogenetic inferences Methodology Methods
Today s topics Inferring phylogeny Introduction! Distance methods! Parsimony method!"#$%&'(!)* +,-.'/01!23454(6!7!2845*0&4'9#6!:&454(6 ;?@AB=C?DEF Overview of phylogenetic inferences Methodology Methods
Hierarchical Clustering
 Hierarchical Clustering Some slides by Serafim Batzoglou 1 From expression profiles to distances From the Raw Data matrix we compute the similarity matrix S. S ij reflects the similarity of the expression
Hierarchical Clustering Some slides by Serafim Batzoglou 1 From expression profiles to distances From the Raw Data matrix we compute the similarity matrix S. S ij reflects the similarity of the expression
 Tree of Life iological Sequence nalysis Chapter http://tolweb.org/tree/ Phylogenetic Prediction ll organisms on Earth have a common ancestor. ll species are related. The relationship is called a phylogeny
Tree of Life iological Sequence nalysis Chapter http://tolweb.org/tree/ Phylogenetic Prediction ll organisms on Earth have a common ancestor. ll species are related. The relationship is called a phylogeny
Estimating Evolutionary Trees. Phylogenetic Methods
 Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
Warm-Up- Review Natural Selection and Reproduction for quiz today!!!! Notes on Evidence of Evolution Work on Vocabulary and Lab
 Date: Agenda Warm-Up- Review Natural Selection and Reproduction for quiz today!!!! Notes on Evidence of Evolution Work on Vocabulary and Lab Ask questions based on 5.1 and 5.2 Quiz on 5.1 and 5.2 How
Date: Agenda Warm-Up- Review Natural Selection and Reproduction for quiz today!!!! Notes on Evidence of Evolution Work on Vocabulary and Lab Ask questions based on 5.1 and 5.2 Quiz on 5.1 and 5.2 How
Constructing Evolutionary Trees
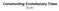 Constructing Evolutionary Trees 0-0 HIV Evolutionary Tree SIVs (monkeys)! HIV (human)! human infection! human HIV/M human HIV/M chimpanzee SIV chimpanzee SIV human HIV/N human HIV/N chimpanzee SIV chimpanzee
Constructing Evolutionary Trees 0-0 HIV Evolutionary Tree SIVs (monkeys)! HIV (human)! human infection! human HIV/M human HIV/M chimpanzee SIV chimpanzee SIV human HIV/N human HIV/N chimpanzee SIV chimpanzee
Methods for solving recurrences
 Methods for solving recurrences Analyzing the complexity of mergesort The merge function Consider the following implementation: 1 int merge ( int v1, int n1, int v, int n ) { 3 int r = malloc ( ( n1+n
Methods for solving recurrences Analyzing the complexity of mergesort The merge function Consider the following implementation: 1 int merge ( int v1, int n1, int v, int n ) { 3 int r = malloc ( ( n1+n
Tree-average distances on certain phylogenetic networks have their weights uniquely determined
 Tree-average distances on certain phylogenetic networks have their weights uniquely determined Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu
Tree-average distances on certain phylogenetic networks have their weights uniquely determined Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu
Phylogeny and Evolution. Gina Cannarozzi ETH Zurich Institute of Computational Science
 Phylogeny and Evolution Gina Cannarozzi ETH Zurich Institute of Computational Science History Aristotle (384-322 BC) classified animals. He found that dolphins do not belong to the fish but to the mammals.
Phylogeny and Evolution Gina Cannarozzi ETH Zurich Institute of Computational Science History Aristotle (384-322 BC) classified animals. He found that dolphins do not belong to the fish but to the mammals.
Phylogenetics: Building Phylogenetic Trees
 1 Phylogenetics: Building Phylogenetic Trees COMP 571 Luay Nakhleh, Rice University 2 Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary model should
1 Phylogenetics: Building Phylogenetic Trees COMP 571 Luay Nakhleh, Rice University 2 Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary model should
Evolutionary trees. Describe the relationship between objects, e.g. species or genes
 Evolutionary trees Bonobo Chimpanzee Human Neanderthal Gorilla Orangutan Describe the relationship between objects, e.g. species or genes Early evolutionary studies Anatomical features were the dominant
Evolutionary trees Bonobo Chimpanzee Human Neanderthal Gorilla Orangutan Describe the relationship between objects, e.g. species or genes Early evolutionary studies Anatomical features were the dominant
Dictionary: an abstract data type
 2-3 Trees 1 Dictionary: an abstract data type A container that maps keys to values Dictionary operations Insert Search Delete Several possible implementations Balanced search trees Hash tables 2 2-3 trees
2-3 Trees 1 Dictionary: an abstract data type A container that maps keys to values Dictionary operations Insert Search Delete Several possible implementations Balanced search trees Hash tables 2 2-3 trees
Assignment 5: Solutions
 Comp 21: Algorithms and Data Structures Assignment : Solutions 1. Heaps. (a) First we remove the minimum key 1 (which we know is located at the root of the heap). We then replace it by the key in the position
Comp 21: Algorithms and Data Structures Assignment : Solutions 1. Heaps. (a) First we remove the minimum key 1 (which we know is located at the root of the heap). We then replace it by the key in the position
Data Structures and Algorithms " Search Trees!!
 Data Structures and Algorithms " Search Trees!! Outline" Binary Search Trees! AVL Trees! (2,4) Trees! 2 Binary Search Trees! "! < 6 2 > 1 4 = 8 9 Ordered Dictionaries" Keys are assumed to come from a total
Data Structures and Algorithms " Search Trees!! Outline" Binary Search Trees! AVL Trees! (2,4) Trees! 2 Binary Search Trees! "! < 6 2 > 1 4 = 8 9 Ordered Dictionaries" Keys are assumed to come from a total
Phylogenetics: Building Phylogenetic Trees. COMP Fall 2010 Luay Nakhleh, Rice University
 Phylogenetics: Building Phylogenetic Trees COMP 571 - Fall 2010 Luay Nakhleh, Rice University Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary
Phylogenetics: Building Phylogenetic Trees COMP 571 - Fall 2010 Luay Nakhleh, Rice University Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary
Dictionary: an abstract data type
 2-3 Trees 1 Dictionary: an abstract data type A container that maps keys to values Dictionary operations Insert Search Delete Several possible implementations Balanced search trees Hash tables 2 2-3 trees
2-3 Trees 1 Dictionary: an abstract data type A container that maps keys to values Dictionary operations Insert Search Delete Several possible implementations Balanced search trees Hash tables 2 2-3 trees
Algorithmic Methods Well-defined methodology Tree reconstruction those that are well-defined enough to be carried out by a computer. Felsenstein 2004,
 Tracing the Evolution of Numerical Phylogenetics: History, Philosophy, and Significance Adam W. Ferguson Phylogenetic Systematics 26 January 2009 Inferring Phylogenies Historical endeavor Darwin- 1837
Tracing the Evolution of Numerical Phylogenetics: History, Philosophy, and Significance Adam W. Ferguson Phylogenetic Systematics 26 January 2009 Inferring Phylogenies Historical endeavor Darwin- 1837
THE THEORY OF EVOLUTION. Darwin, the people who contributed to his ideas, and what it all really means.
 THE THEORY OF EVOLUTION Darwin, the people who contributed to his ideas, and what it all really means. DARWIN S JOURNEY Charles Darwin was born in England on February 12, 1809. Geologists were suggesting
THE THEORY OF EVOLUTION Darwin, the people who contributed to his ideas, and what it all really means. DARWIN S JOURNEY Charles Darwin was born in England on February 12, 1809. Geologists were suggesting
Statistical inference provides methods for drawing conclusions about a population from sample data.
 Introduction to inference Confidence Intervals Statistical inference provides methods for drawing conclusions about a population from sample data. 10.1 Estimating with confidence SAT σ = 100 n = 500 µ
Introduction to inference Confidence Intervals Statistical inference provides methods for drawing conclusions about a population from sample data. 10.1 Estimating with confidence SAT σ = 100 n = 500 µ
Advanced Analysis of Algorithms - Midterm (Solutions)
 Advanced Analysis of Algorithms - Midterm (Solutions) K. Subramani LCSEE, West Virginia University, Morgantown, WV {ksmani@csee.wvu.edu} 1 Problems 1. Solve the following recurrence using substitution:
Advanced Analysis of Algorithms - Midterm (Solutions) K. Subramani LCSEE, West Virginia University, Morgantown, WV {ksmani@csee.wvu.edu} 1 Problems 1. Solve the following recurrence using substitution:
Phylogeny: traditional and Bayesian approaches
 Phylogeny: traditional and Bayesian approaches 5-Feb-2014 DEKM book Notes from Dr. B. John Holder and Lewis, Nature Reviews Genetics 4, 275-284, 2003 1 Phylogeny A graph depicting the ancestor-descendent
Phylogeny: traditional and Bayesian approaches 5-Feb-2014 DEKM book Notes from Dr. B. John Holder and Lewis, Nature Reviews Genetics 4, 275-284, 2003 1 Phylogeny A graph depicting the ancestor-descendent
Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction. Lesser Tenrec (Echinops telfairi)
 Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction Lesser Tenrec (Echinops telfairi) Goals: 1. Use phylogenetic experimental design theory to select optimal taxa to
Phylogenetics - Orthology, phylogenetic experimental design and phylogeny reconstruction Lesser Tenrec (Echinops telfairi) Goals: 1. Use phylogenetic experimental design theory to select optimal taxa to
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut
 Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
9/30/11. Evolution theory. Phylogenetic Tree Reconstruction. Phylogenetic trees (binary trees) Phylogeny (phylogenetic tree)
 I9 Introduction to Bioinformatics, 0 Phylogenetic ree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & omputing, IUB Evolution theory Speciation Evolution of new organisms is driven by
I9 Introduction to Bioinformatics, 0 Phylogenetic ree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & omputing, IUB Evolution theory Speciation Evolution of new organisms is driven by
Molecular phylogeny - Using molecular sequences to infer evolutionary relationships. Tore Samuelsson Feb 2016
 Molecular phylogeny - Using molecular sequences to infer evolutionary relationships Tore Samuelsson Feb 2016 Molecular phylogeny is being used in the identification and characterization of new pathogens,
Molecular phylogeny - Using molecular sequences to infer evolutionary relationships Tore Samuelsson Feb 2016 Molecular phylogeny is being used in the identification and characterization of new pathogens,
week: 4 Date: Microscopes Cell Structure Cell Function Standards None 1b, 1h 1b, 1h, 4f, 5a 1a, 1c, 1d, 1e, 1g, 1j
 July, 2004 week: 1 Topics Course introduction Lab Safety week: 2 Introduction to chemistry Chapter summarizing Note Taking week: 3 Biochemistry: Compounds of life week: 4 Microscopes Cell Structure Cell
July, 2004 week: 1 Topics Course introduction Lab Safety week: 2 Introduction to chemistry Chapter summarizing Note Taking week: 3 Biochemistry: Compounds of life week: 4 Microscopes Cell Structure Cell
Using phylogenetics to estimate species divergence times... Basics and basic issues for Bayesian inference of divergence times (plus some digression)
 Using phylogenetics to estimate species divergence times... More accurately... Basics and basic issues for Bayesian inference of divergence times (plus some digression) "A comparison of the structures
Using phylogenetics to estimate species divergence times... More accurately... Basics and basic issues for Bayesian inference of divergence times (plus some digression) "A comparison of the structures
Darwin & Evolution by Natural Selection
 Darwin & Evolution by Natural Selection Warbler Cactus eater Insect eaters Seed eaters Bud eater 2006-2007 Charles Darwin Proposed a way how evolution works u How did creatures change over time? u by natural
Darwin & Evolution by Natural Selection Warbler Cactus eater Insect eaters Seed eaters Bud eater 2006-2007 Charles Darwin Proposed a way how evolution works u How did creatures change over time? u by natural
Building Phylogenetic Trees UPGMA & NJ
 uilding Phylogenetic Trees UPGM & NJ UPGM UPGM Unweighted Pair-Group Method with rithmetic mean Unweighted = all pairwise distances contribute equally. Pair-Group = groups are combined in pairs. rithmetic
uilding Phylogenetic Trees UPGM & NJ UPGM UPGM Unweighted Pair-Group Method with rithmetic mean Unweighted = all pairwise distances contribute equally. Pair-Group = groups are combined in pairs. rithmetic
Evidences of Evolution (Clues)
 Evidences of Evolution (Clues) Darwin stated that all organisms descended from a common ancestor Darwin based his theory of Natural Selection on observations of: Traits, geographical distribution, selective
Evidences of Evolution (Clues) Darwin stated that all organisms descended from a common ancestor Darwin based his theory of Natural Selection on observations of: Traits, geographical distribution, selective
Skip Lists. What is a Skip List. Skip Lists 3/19/14
 Presentation for use with the textbook Data Structures and Algorithms in Java, 6 th edition, by M. T. Goodrich, R. Tamassia, and M. H. Goldwasser, Wiley, 2014 Skip Lists 15 15 23 10 15 23 36 Skip Lists
Presentation for use with the textbook Data Structures and Algorithms in Java, 6 th edition, by M. T. Goodrich, R. Tamassia, and M. H. Goldwasser, Wiley, 2014 Skip Lists 15 15 23 10 15 23 36 Skip Lists
SEVENTH EDITION and EXPANDED SEVENTH EDITION
 SEVENTH EDITION and EXPANDED SEVENTH EDITION Slide 10-1 Chapter 10 Mathematical Systems 10.1 Groups Definitions A mathematical system consists of a set of elements and at least one binary operation. A
SEVENTH EDITION and EXPANDED SEVENTH EDITION Slide 10-1 Chapter 10 Mathematical Systems 10.1 Groups Definitions A mathematical system consists of a set of elements and at least one binary operation. A
How to read and make phylogenetic trees Zuzana Starostová
 How to read and make phylogenetic trees Zuzana Starostová How to make phylogenetic trees? Workflow: obtain DNA sequence quality check sequence alignment calculating genetic distances phylogeny estimation
How to read and make phylogenetic trees Zuzana Starostová How to make phylogenetic trees? Workflow: obtain DNA sequence quality check sequence alignment calculating genetic distances phylogeny estimation
Properties of normal phylogenetic networks
 Properties of normal phylogenetic networks Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu August 13, 2009 Abstract. A phylogenetic network is
Properties of normal phylogenetic networks Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu August 13, 2009 Abstract. A phylogenetic network is
Evolution of Body Size in Bears. How to Build and Use a Phylogeny
 Evolution of Body Size in Bears How to Build and Use a Phylogeny Lesson II Using a Phylogeny Objectives for Lesson II: 1. Overview of concepts 1. Simple ancestral state reconstruction on the bear phylogeny
Evolution of Body Size in Bears How to Build and Use a Phylogeny Lesson II Using a Phylogeny Objectives for Lesson II: 1. Overview of concepts 1. Simple ancestral state reconstruction on the bear phylogeny
How should we organize the diversity of animal life?
 How should we organize the diversity of animal life? The difference between Taxonomy Linneaus, and Cladistics Darwin What are phylogenies? How do we read them? How do we estimate them? Classification (Taxonomy)
How should we organize the diversity of animal life? The difference between Taxonomy Linneaus, and Cladistics Darwin What are phylogenies? How do we read them? How do we estimate them? Classification (Taxonomy)
Incremental Phylogenetics by Repeated Insertions: An Evolutionary Tree Algorithm
 Incremental Phylogenetics by Repeated Insertions: An Evolutionary Tree Algorithm Peter Z. Revesz, Zhiqiang Li Abstract We introduce the idea of constructing hypothetical evolutionary trees using an incremental
Incremental Phylogenetics by Repeated Insertions: An Evolutionary Tree Algorithm Peter Z. Revesz, Zhiqiang Li Abstract We introduce the idea of constructing hypothetical evolutionary trees using an incremental
Warm Up. Explain how a mutation can be detrimental in one environmental context and beneficial in another.
 Warm Up Explain how a mutation can be detrimental in one environmental context and beneficial in another. Last Picture 4B Evidence for Evolution 1A.4a: Scientific evidence of biological evolution uses
Warm Up Explain how a mutation can be detrimental in one environmental context and beneficial in another. Last Picture 4B Evidence for Evolution 1A.4a: Scientific evidence of biological evolution uses
Phylogenetics. BIOL 7711 Computational Bioscience
 Consortium for Comparative Genomics! University of Colorado School of Medicine Phylogenetics BIOL 7711 Computational Bioscience Biochemistry and Molecular Genetics Computational Bioscience Program Consortium
Consortium for Comparative Genomics! University of Colorado School of Medicine Phylogenetics BIOL 7711 Computational Bioscience Biochemistry and Molecular Genetics Computational Bioscience Program Consortium
BIOL 1010 Introduction to Biology: The Evolution and Diversity of Life. Spring 2011 Sections A & B
 BIOL 1010 Introduction to Biology: The Evolution and Diversity of Life. Spring 2011 Sections A & B Steve Thompson: stthompson@valdosta.edu http://www.bioinfo4u.net 1 ʻTree of Life,ʼ ʻprimitive,ʼ ʻprogressʼ
BIOL 1010 Introduction to Biology: The Evolution and Diversity of Life. Spring 2011 Sections A & B Steve Thompson: stthompson@valdosta.edu http://www.bioinfo4u.net 1 ʻTree of Life,ʼ ʻprimitive,ʼ ʻprogressʼ
Phylogenetic analyses. Kirsi Kostamo
 Phylogenetic analyses Kirsi Kostamo The aim: To construct a visual representation (a tree) to describe the assumed evolution occurring between and among different groups (individuals, populations, species,
Phylogenetic analyses Kirsi Kostamo The aim: To construct a visual representation (a tree) to describe the assumed evolution occurring between and among different groups (individuals, populations, species,
Change Over Time Concept Map
 Change Over Time Concept Map Darwin reasoned that plants or animals that arrived on the Galapagos Islands faced conditions that were different from those on the mainland. Perhaps, Darwin hypothesized,
Change Over Time Concept Map Darwin reasoned that plants or animals that arrived on the Galapagos Islands faced conditions that were different from those on the mainland. Perhaps, Darwin hypothesized,
Lecture 6 Phylogenetic Inference
 Lecture 6 Phylogenetic Inference From Darwin s notebook in 1837 Charles Darwin Willi Hennig From The Origin in 1859 Cladistics Phylogenetic inference Willi Hennig, Cladistics 1. Clade, Monophyletic group,
Lecture 6 Phylogenetic Inference From Darwin s notebook in 1837 Charles Darwin Willi Hennig From The Origin in 1859 Cladistics Phylogenetic inference Willi Hennig, Cladistics 1. Clade, Monophyletic group,
molecular evolution and phylogenetics
 molecular evolution and phylogenetics Charlotte Darby Computational Genomics: Applied Comparative Genomics 2.13.18 https://www.thinglink.com/scene/762084640000311296 Internal node Root TIME Branch Leaves
molecular evolution and phylogenetics Charlotte Darby Computational Genomics: Applied Comparative Genomics 2.13.18 https://www.thinglink.com/scene/762084640000311296 Internal node Root TIME Branch Leaves
Biology 1B Evolution Lecture 2 (February 26, 2010) Natural Selection, Phylogenies
 1 Natural Selection (Darwin-Wallace): There are three conditions for natural selection: 1. Variation: Individuals within a population have different characteristics/traits (or phenotypes). 2. Inheritance:
1 Natural Selection (Darwin-Wallace): There are three conditions for natural selection: 1. Variation: Individuals within a population have different characteristics/traits (or phenotypes). 2. Inheritance:
Molecular Phylogenetics (part 1 of 2) Computational Biology Course João André Carriço
 Molecular Phylogenetics (part 1 of 2) Computational Biology Course João André Carriço jcarrico@fm.ul.pt Charles Darwin (1809-1882) Charles Darwin s tree of life in Notebook B, 1837-1838 Ernst Haeckel (1934-1919)
Molecular Phylogenetics (part 1 of 2) Computational Biology Course João André Carriço jcarrico@fm.ul.pt Charles Darwin (1809-1882) Charles Darwin s tree of life in Notebook B, 1837-1838 Ernst Haeckel (1934-1919)
Bioinformatics 1. Sepp Hochreiter. Biology, Sequences, Phylogenetics Part 4. Bioinformatics 1: Biology, Sequences, Phylogenetics
 Bioinformatics 1 Biology, Sequences, Phylogenetics Part 4 Sepp Hochreiter Klausur Mo. 30.01.2011 Zeit: 15:30 17:00 Raum: HS14 Anmeldung Kusss Contents Methods and Bootstrapping of Maximum Methods Methods
Bioinformatics 1 Biology, Sequences, Phylogenetics Part 4 Sepp Hochreiter Klausur Mo. 30.01.2011 Zeit: 15:30 17:00 Raum: HS14 Anmeldung Kusss Contents Methods and Bootstrapping of Maximum Methods Methods
At least one of us is a knave. What are A and B?
 1. This is a puzzle about an island in which everyone is either a knight or a knave. Knights always tell the truth and knaves always lie. This problem is about two people A and B, each of whom is either
1. This is a puzzle about an island in which everyone is either a knight or a knave. Knights always tell the truth and knaves always lie. This problem is about two people A and B, each of whom is either
Reconstruire le passé biologique modèles, méthodes, performances, limites
 Reconstruire le passé biologique modèles, méthodes, performances, limites Olivier Gascuel Centre de Bioinformatique, Biostatistique et Biologie Intégrative C3BI USR 3756 Institut Pasteur & CNRS Reconstruire
Reconstruire le passé biologique modèles, méthodes, performances, limites Olivier Gascuel Centre de Bioinformatique, Biostatistique et Biologie Intégrative C3BI USR 3756 Institut Pasteur & CNRS Reconstruire
Properties of Life. Levels of Organization. Levels of Organization. Levels of Organization. Levels of Organization. The Science of Biology.
 The Science of Biology Chapter 1 Properties of Life Living organisms: are composed of cells are complex and ordered respond to their environment can grow and reproduce obtain and use energy maintain internal
The Science of Biology Chapter 1 Properties of Life Living organisms: are composed of cells are complex and ordered respond to their environment can grow and reproduce obtain and use energy maintain internal
2.1 Computational Tractability. Chapter 2. Basics of Algorithm Analysis. Computational Tractability. Polynomial-Time
 Chapter 2 2.1 Computational Tractability Basics of Algorithm Analysis "For me, great algorithms are the poetry of computation. Just like verse, they can be terse, allusive, dense, and even mysterious.
Chapter 2 2.1 Computational Tractability Basics of Algorithm Analysis "For me, great algorithms are the poetry of computation. Just like verse, they can be terse, allusive, dense, and even mysterious.
2. ALGORITHM ANALYSIS
 2. ALGORITHM ANALYSIS computational tractability asymptotic order of growth survey of common running times Lecture slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley http://www.cs.princeton.edu/~wayne/kleinberg-tardos
2. ALGORITHM ANALYSIS computational tractability asymptotic order of growth survey of common running times Lecture slides by Kevin Wayne Copyright 2005 Pearson-Addison Wesley http://www.cs.princeton.edu/~wayne/kleinberg-tardos
Inferring phylogeny. Constructing phylogenetic trees. Tõnu Margus. Bioinformatics MTAT
 Inferring phylogeny Constructing phylogenetic trees Tõnu Margus Contents What is phylogeny? How/why it is possible to infer it? Representing evolutionary relationships on trees What type questions questions
Inferring phylogeny Constructing phylogenetic trees Tõnu Margus Contents What is phylogeny? How/why it is possible to infer it? Representing evolutionary relationships on trees What type questions questions
Reading for Lecture 13 Release v10
 Reading for Lecture 13 Release v10 Christopher Lee November 15, 2011 Contents 1 Evolutionary Trees i 1.1 Evolution as a Markov Process...................................... ii 1.2 Rooted vs. Unrooted Trees........................................
Reading for Lecture 13 Release v10 Christopher Lee November 15, 2011 Contents 1 Evolutionary Trees i 1.1 Evolution as a Markov Process...................................... ii 1.2 Rooted vs. Unrooted Trees........................................
Consistency Index (CI)
 Consistency Index (CI) minimum number of changes divided by the number required on the tree. CI=1 if there is no homoplasy negatively correlated with the number of species sampled Retention Index (RI)
Consistency Index (CI) minimum number of changes divided by the number required on the tree. CI=1 if there is no homoplasy negatively correlated with the number of species sampled Retention Index (RI)
Session 5: Phylogenomics
 Session 5: Phylogenomics B.- Phylogeny based orthology assignment REMINDER: Gene tree reconstruction is divided in three steps: homology search, multiple sequence alignment and model selection plus tree
Session 5: Phylogenomics B.- Phylogeny based orthology assignment REMINDER: Gene tree reconstruction is divided in three steps: homology search, multiple sequence alignment and model selection plus tree
Comparative Genomics II
 Comparative Genomics II Advances in Bioinformatics and Genomics GEN 240B Jason Stajich May 19 Comparative Genomics II Slide 1/31 Outline Introduction Gene Families Pairwise Methods Phylogenetic Methods
Comparative Genomics II Advances in Bioinformatics and Genomics GEN 240B Jason Stajich May 19 Comparative Genomics II Slide 1/31 Outline Introduction Gene Families Pairwise Methods Phylogenetic Methods
Fig. 26.7a. Biodiversity. 1. Course Outline Outcomes Instructors Text Grading. 2. Course Syllabus. Fig. 26.7b Table
 Fig. 26.7a Biodiversity 1. Course Outline Outcomes Instructors Text Grading 2. Course Syllabus Fig. 26.7b Table 26.2-1 1 Table 26.2-2 Outline: Systematics and the Phylogenetic Revolution I. Naming and
Fig. 26.7a Biodiversity 1. Course Outline Outcomes Instructors Text Grading 2. Course Syllabus Fig. 26.7b Table 26.2-1 1 Table 26.2-2 Outline: Systematics and the Phylogenetic Revolution I. Naming and
