c 2004 Society for Industrial and Applied Mathematics
|
|
|
- Leonard Hopkins
- 5 years ago
- Views:
Transcription
1 SIAM J. COMPUT. Vol. 33, No. 3, pp c 2004 Society for Industrial and Applied Mathematics INCOMPLETE DIRECTED PERFECT PHYLOGENY ITSIK PE ER, TAL PUPKO, RON SHAMIR, AND RODED SHARAN Abstract. Perfect phylogeny is one of the fundamental models for studying evolution. We investigate the following variant of the model: The input is a species-characters matrix. The characters are binary and directed; i.e., a species can only gain characters. The difference from standard perfect phylogeny is that for some species the states of some characters are unknown. The question is whether one can complete the missing states in a way that admits a perfect phylogeny. The problem arises in classical phylogenetic studies, when some states are missing or undetermined. Quite recently, studies that infer phylogenies using inserted repeat elements in DNA gave rise to the same problem. Extant solutions for it take time O(n 2 m) for n species and m characters. We provide a graph theoretic formulation of the problem as a graph sandwich problem, and give near-optimal Õ(nm)-time algorithms for the problem. We also study the problem of finding a single, general solution tree, from which any other solution can be obtained by node splitting. We provide an algorithm to construct such a tree, or determine that none exists. Key words. perfect phylogeny, incomplete data, graph sandwich, evolution AMS subject classifications. 05C85, 05C50, 92D15 DOI /S Introduction. When studying evolution, the divergence patterns leading from a single ancestor species to its contemporary descendants are usually modeled by a tree structure, called phylogenetic tree, orphylogeny. Extant species correspond to the tree leaves, while their common progenitor corresponds to the root. Internal nodes correspond to hypothetical ancestral species, which putatively split up and evolved into distinct species. Tree branches model changes through time of the hypothetical ancestor species. The common case is that one has information regarding the leaves, from which the phylogenetic tree is to be inferred. This task, called phylogenetic reconstruction (cf. [8]), was one of the first algorithmic challenges posed by biology, and the computational community has been dealing with problems of this flavor for over three decades (see, e.g., [13]). The character-based approach to tree reconstruction describes extant species by their attributes or characters. Each character takes on one of several possible states. The input is represented by a matrix A, where a ij is the state of character j in Received by the editors April 29, 2002; accepted for publication (in revised form) December 19, 2003; published electronically March 23, Portions of this paper appeared as Incomplete directed perfect phylogeny, in Proceedings of the Eleventh Annual Symposium on Combinatorial Pattern Matching, Lecture Notes in Comput. Sci. 1848, Springer-Verlag, Berlin, 2000, pp Other parts appeared as On the generality of phylogenies from incomplete directed characters, in Proceedings of the Eighth Scandinavian Workshop on Algorithm Theory, Lecture Notes in Comput. Sci. 2368, Springer-Verlag, New York, 2002, pp Medical and Population Genetics Group, Broad Institute, 9 Cambridge Center, Cambridge, MA (peer@broad.mit.edu). This author s research was supported by a Clore foundation scholarship. Department of Cell Research and Immunology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv 69978, Israel (talp@post.tau.ac.il). School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel (rshamir@tau.ac.il). This author s research was supported in part by a grant from the Israel Science Foundation (grant 309/02). Corresponding author. International Computer Science Institute, 1947 Center St., Berkeley, CA (roded@icsi.berkeley.edu). This author s research was supported by a Fulbright grant and by an Eshkol fellowship from the Ministry of Science, Israel. 590
2 INCOMPLETE DIRECTED PERFECT PHYLOGENY 591 species i, and the ith row is the character vector of species i. The output sought is a hypothesis regarding evolution, i.e., a phylogenetic tree along with the suggested character vectors of the internal nodes. This output must satisfy properties specified by the problem variant. One important class of phylogenetic reconstruction problems concerns finding a perfect phylogeny. The property required from such a phylogeny is that for each possible character state, the set of all nodes that have that state induces a connected subtree. The general perfect phylogeny problem is NP-hard [5, 21]. When considering the number of possible states per character as a parameter, the problem is fixed parameter tractable [2, 16]. For binary characters, having only two possible states, perfect phylogeny is linear-time solvable [12]. When no perfect phylogeny is possible, one option is to seek a largest subset of characters which admits a perfect phylogeny. Characters in such a subset are said to be compatible. Compatibility problems have been studied extensively (see, e.g., [18]). Another common optimization approach to phylogenetic reconstruction is the parsimony criterion. It calls for a solution with the fewest state changes altogether, counting a change whenever the state of a character changes between a species and its ancestor species. This problem is known to be NP-hard [9]. A variant introduced by Camin and Sokal [6] assumes that characters are binary and directed, namely, only 0 1 changes may occur on any path from the root to a leaf. Denoting by 1 and 0 the presence and absence, respectively, of the character, this means that characters can only be gained throughout evolution. Another related binary variant is Dollo parsimony [7, 20], which assumes that a 0 1 change may happen only once; i.e., a character can be gained once, but it can be lost several times. Both of these variants are polynomially solvable (cf. [8]). In this paper, we discuss a variant of binary perfect phylogeny which combines assumptions of both Camin Sokal parsimony and Dollo parsimony. The setup is as follows: The characters are binary, directed, and can be gained only once. As in perfect phylogeny, the input is a matrix of character vectors, with the difference that some character states are missing. The question is whether one can complete the missing states in a way admitting a perfect phylogeny. We call this problem Incomplete Directed Perfect phylogeny (IDP). The problem of handling incomplete phylogenetic data arises whenever some of the data are missing. It is also encountered in the context of morphological characters, where for some species it may be impossible to reliably assign a state to a character. The problem of determining whether a set of incomplete undirected characters is compatible was shown to be NP-complete, even in the case of binary characters [21]. Indeed, the popular PAUP software package [22] provides an exponential solution to the problem by exhaustively searching the space of missing states. Quite recently, a novel kind of genomic data has given rise to the same problem: Nikaido, Rooney, and Okado [19] use inserted repetitive genomic elements, particularly Short Interspersed Nuclear Elements (SINEs), as a source of evolutionary information. SINEs are short DNA sequences that were copied and randomly reinserted into various genomic loci during evolution. The distinct insertion loci are identifiable by the flanking sequences on both sides of the insertion site (see Figure 1). These insertions are assumed to be unique events in evolution, because the odds of having separate insertion events at the very same locus are negligible. Furthermore, a SINE insertion is assumed to be irreversible; i.e., once a SINE sequence has been inserted somewhere along the genome, it is practically impossible for the exact, complete SINE to leave
3 592 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN Genome 1 Legend: SINE Genome 2 Locus A Locus B Genome 3 Locus C Fig. 1. SINEs (black boxes) repeat in different loci (different shades of grey) across distinct genomes. A SINE insertion transformed Genome 1 into Genome 2. A deletion of a locus transformed Genome 2 into Genome 3. Given Genomes 1 and 3, we can identify that the SINE on locus C is not present in Genome 1, by its flanking sequence. However, locus B is missing in Genome 3. that specific locus. However, the inserted segment along with its flanking sequences may be lost when a large genomic region, which includes them, is deleted. In that case we do not know whether a SINE insertion had occurred in the missing site prior to its deletion. One can model such data by assigning to each locus a character, whose state is 1 if the SINE occurred in that locus, 0 if the locus is present but does not contain the SINE, and? if the locus is missing. The resulting reconstruction problem is precisely IDP. The IDP problem becomes polynomial when the characters are directed: Benham et al. [4] studied the compatibility problem on generalized characters. Their work implies an O(n 2 m)-time algorithm for IDP, where n and m denote the number of species and characters, respectively. Another problem related to IDP is the consensus tree problem [3, 14]. This problem calls for constructing a consensus tree from binary subtrees, and is solvable in polynomial time. One can reduce IDP to the latter problem, but the reduction itself takes Ω(n 2 m) time. Our approach to the IDP problem is graph theoretic. We first provide several graph and matrix characterizations for solvable instances of binary directed perfect phylogeny. We then reformulate IDP as a graph sandwich problem: The input data is recast as two nested graphs, and solving IDP is shown to be equivalent to finding a graph of a particular type sandwiched between them. This formulation allows us to devise efficient algorithms for IDP. We provide two algorithms for IDP, which we call Algorithms A and B. Algorithm A has two possible implementations: deterministic and randomized. Its deterministic complexity is O(nm + k log 2 (n + m)), for an instance with k 1-entries in the species-characters matrix. The randomized version of Algorithm A takes O(nm + k log(l 2 /k)+l(log l) 3 log log l) expected time, where l = n + m. Algorithm B is deterministic and takes O(l 2 log l) time. For both algorithms, the improved complexity is obtained by using dynamic data structures for maintaining the connected components of a graph [23, 14, 15]. Since an Ω(nm) lower bound was shown by Gusfield for directed binary perfect phylogeny [12], our algorithms have near-optimal time complexity. We also study the issue of multiple solutions for IDP. Often there is more than one phylogeny that is consistent with the data. When the input matrix is complete and has a solution, there is always a tree T that is general; i.e., it is a solution, and every other tree consistent with the data can be obtained from T by node splitting. In other words, T describes all the definite information in the data, and ambiguities (nodes
4 INCOMPLETE DIRECTED PERFECT PHYLOGENY 593 with three or more children) can be resolved by additional information. This is not always the case if the data matrix is incomplete: There may or may not be a general solution tree. In that case, using a particular solution and additional information, one can conclude that the data is inconsistent, even though the additional information may be consistent with another solution. It is thus desirable to know if a general solution exists and to generate such a solution if the answer is positive. We provide answers to both questions. We prove that Algorithm A provides the general solution of a problem instance, if such exists. We also give an algorithm which determines if the solution T produced by Algorithm A is indeed general. The complexity of the latter algorithm is O(nm + kd), where d denotes the maximum out-degree of T. The paper is organized as follows. In section 2 we provide some preliminaries, and formalize the IDP problem. In section 3 we characterize binary matrices admitting a directed perfect phylogeny, and provide the graph sandwich formulation for IDP. In section 4 we present algorithms for IDP. Finally in section 5 we analyze the generality of the solution produced by Algorithm A. 2. Preliminaries. We first specify some terminology and notation. We reserve the terms nodes and branches for trees, and use the terms vertices and edges for other graphs. Matrices are denoted by an upper-case letter, while their elements are denoted by the corresponding lower-case letter. Let G =(V,E) be a graph. We denote its set of vertices also by V (G), and its set of edges also by E(G). Let V V be a subset of the vertices. The subgraph induced by V is the graph (V,E (V V )). We say that V is connected in G, if V is contained in some connected component of G. The length of a path in G is the number of edges along it. Let T be a rooted tree with leaf set S, where branches are directed from the root towards the leaves. The out-degree of a node x in T is its number of children, and is denoted by d(x). Foranodexin T we denote the leaf set of the subtree rooted at x by L(x). L(x) is called a clade of T. For consistency, we consider to be a clade of T as well, and call it the empty clade. S,, and all singletons are called trivial clades. We denote by triv(s) the collection of all trivial clades. Two sets are said to be compatible if they are either disjoint, or one of them contains the other. Observation 1 (cf. [18]). A collection S of subsets of a set S is the set of clades of some tree over S if and only if S contains triv(s) and its subsets are pairwise compatible. A tree T is uniquely characterized by its set of clades. The transformation between a branch-node representation of a tree and a list of its clades is straightforward. Thus, we hereafter identify a tree with the set of its clades, and use the notation S T to indicate that S is a clade of T.IfŜ is a subset of the leaves of T, then the subtree of T induced on Ŝ is the collection {Ŝ S : S T}(which defines a tree). Throughout the paper we denote by S = {s 1,...,s n } the set of all species and by C = {c 1,...,c m } the set of all (binary) characters. For a graph K, we define C(K) C V (K) and S(K) S V (K). Let B n m be a binary matrix whose rows correspond to species, each row being the character vector of its corresponding species. That is, b ij = 1 if and only if the species s i has the character c j.aphylogenetic tree for B is a rooted tree T with n leaves corresponding to the n species of S, such that each character is associated with a clade S of T, and the following properties are satisfied: (1) If c j is associated with S, then s i S if and only if b ij =1.
5 594 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN Characters c 1 c 2 c 3 c 4 c 5 c 1 1? s Species?? s c 5? 0 1?? s c 2,c 4 c 3 s 2 s 1 s 3 Fig. 2. Left to right: An incomplete matrix A, a completion B of A, and a phylogenetic tree that explains A via B. Each character is written to the right of its origin node. Characters Species? 1 1 0? 1 Fig. 3. An incomplete matrix which has no phylogenetic tree although every pair of its columns has one. (2) Every nontrivial clade of T is associated with at least one character. For a character c, the node x of T whose clade L(x) is associated with c is called the origin of c with respect to T. Characters associated with have no origin. A {0, 1,?} matrix is called incomplete. For convenience, we consider binary matrices as incomplete. Let A n m be a {0, 1,?} matrix in which a ij =1ifs i has c j, a ij =0ifs i lacks c j, and a ij =? if it is not known whether s i has c j. For a character c j and a state x {0, 1,?}, the x-set of c j in A is the set of species {s i S : a ij = x}. c j is called a null character if its 1-set is empty. For subsets Ŝ S and Ĉ C, define A Ŝ, Ĉ to be the submatrix of A induced on Ŝ Ĉ. A binary matrix B is called a completion of A if a ij {b ij,?} for all i, j. Thus, a completion replaces all the? s in A by zeroes and ones. If B has a phylogenetic tree T, we say that T is a phylogenetic tree for A as well. We also say that T explains A via B, and that A is explainable. An example of these definitions is given in Figure 2. The following lemma, closely related to Observation 1, has been proved independently by several authors. Lemma 2 (cf. [18]). A binary matrix B has a phylogenetic tree if and only if the 1-sets of every two characters are compatible. An analogous lemma holds for undirected characters (cf. [12]). In contrast, for incomplete matrices, even if every pair of columns has a phylogenetic tree, the full matrix might not have one. An example of such a matrix was provided in [8] for incomplete undirected characters. We provide a simpler example for incomplete directed characters in Figure 3. Indeed, if we consider columns 1 and 2 in the example, then the missing entry on column 1 should be completed to 1 and the one on column 2 should be completed to 0. However, in such a completion the characters on columns 2 and 3 are not compatible.
6 INCOMPLETE DIRECTED PERFECT PHYLOGENY 595 Characters c c Species s s s Fig. 4. The Σ subgraph. We are now ready to state the IDP problem as follows: Incomplete Directed Perfect Phylogeny (IDP): Instance: An incomplete matrix A. Goal: Find a phylogenetic tree for A, or determine that no such tree exists. In section 3 we characterize complete binary matrices that admit a perfect phylogeny. In section 4 we present our algorithmic approaches for IDP. 3. Characterizations of explainable binary matrices Forbidden subgraph characterization. Let B be a species-characters binary matrix of order n m. Construct the bipartite graph G(B) =(S, C, E) with E = {(s i,c j ):b ij =1}. A Σ subgraph is an induced subgraph of G(B) that includes three vertices from S, two vertices from C, and exactly four edges, forming a path of length 4 in G(B) (see Figure 4). A bipartite graph with no induced Σ subgraph is called Σ-free. The following theorem restates Lemma 2 in terms of graph theory. Theorem 3. B has a phylogenetic tree if and only if G(B) is Σ-free. Corollary 4. Let Ŝ S and Ĉ C be subsets of the species and characters, respectively. If A is explainable, then so is A Ŝ, Ĉ. Observation 5. Let A be a matrix explained by a tree T and Let Ŝ = L(x) be a clade in T, where x isanodeoft. Then the submatrix A Ŝ,C is explained by the subtree of T rooted at x. For a subset S S of species, we say that a character c is S -universal in B, if its 1-set (in B) contains S. Proposition 6. If G(B) is connected and Σ-free, then there exists a character which is S-universal in B. Proof. Suppose to the contrary that B has no S-universal character. Consider the collection of all 1-sets of characters in B. Let c be a character whose 1-set is maximal with respect to inclusion in this collection. Let s be a species which lacks c. Since G(B) is connected, there exists a path from s to c in G(B). Consider a shortest such path P. Since G(B) is bipartite, the length of P is odd. However, P cannot be of length 1, by the choice of s. Furthermore, if P is of length greater than 3, then its first five vertices induce a Σ subgraph, a contradiction. Thus P =(s,c,s,c) must be of length 3. By maximality of the 1-set of c, it is not contained in the 1-set of c. Hence, there exists a species s which has the character c but lacks c. Together with s, the vertices of P induce a Σ subgraph, as depicted in Figure 4, a contradiction. Let Ψ be a graph property. In the Ψ sandwich problem one is given a vertex set V and a partition of V V into three disjoint subsets: E 0 forbidden edges, E 1 mandatory edges, and E? optional edges. The objective is to find a supergraph of (V,E 1 ) which satisfies Ψ and contains no forbidden edges. Hence, the required graph (V,F) must be sandwiched between (V,E 1 ) and (V,E 1 E? ). The reader is referred to articles [10, 11] for a discussion of various sandwich problems.
7 596 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN For the property containing no induced Σ subgraph (a property of bipartite graphs) the sandwich problem is defined as follows: Σ-free Sandwich: Instance: A vertex set V = S C with S C =, and a partition E 0 E? E 1 of S C. Goal: Find a set of edges F such that F E 1, F E 0 =, and the graph (V,F) is Σ-free, or determine that no such set exists. Theorem 3 motivates looking at the IDP problem with input A as an instance ((S, C),E0 A,E? A,EA 1 ) of the Σ-free sandwich problem. Here, Ex A = {(s i,c j ):a ij = x} for x =0,?, 1. In what follows, we omit the superscript A when it is clear from the context. Proposition 7. The Σ-free sandwich problem is equivalent to IDP. Note that there is an obvious 1-1 correspondence between completions of A and possible solutions of the corresponding sandwich instance ((S, C),E 0,E?,E 1 ). Hence, in what follows we refer to matrices and their corresponding sandwich instances interchangeably Forbidden submatrix characterizations. A binary matrix B is called good if it can be decomposed as follows: (1) Its left k 1 0 columns are all ones. (2) There exist good matrices B 1,...,B l, such that the rest (0 or more) of the columns of B form the block-structure illustrated in Figure 5. A matrix A is canonical if A =[B, C], where B is a zero submatrix and C is good. We say that a matrix B avoids a matrix X, if no submatrix of B is identical to X. Theorem 8. Let B be a binary matrix. The following are equivalent: 1. B has a phylogenetic tree. 2. G(B) is Σ-free. 3. Every matrix obtained by permuting the rows and columns of B avoids the following matrix: Z = There exists an ordering of the rows and columns of B which yields a canonical matrix. 5. There exists an ordering of the rows and columns of B so that the resulting matrix avoids the following matrices: X 1 = [ ] [ 0 1, X 2 = 1 1. ] [ 1 1, X 3 = 0 1 ], X 4 = The reader is referred to article [17] for other problems of permuting matrices to avoid forbidden submatrices. Proof. 1 2 Theorem Trivial. 1 4 Suppose T is a tree that explains B. Assign to each node of T an index which equals its position in a preorder visit of T. Sort the characters according to the indices of their origin nodes, letting null characters come first. Sort
8 INCOMPLETE DIRECTED PERFECT PHYLOGENY 597 B 1 B B B l Fig. 5. Construction of a good matrix. Each B i is a good matrix. A canonical matrix is formed from it by appending columns of zeroes on the left. the species according to the indices of their corresponding leaves in T. The result is a canonical matrix. 4 5 It is easy to verify that canonical matrices avoid X 1,...,X Suppose to the contrary that B has an ordering of its rows and columns, so that rows i 1,i 2,i 3 and columns j 1,j 2 of the resulting matrix form the submatrix Z. Consider the permutations θ row,θ col of the rows and columns of B, respectively, which yield a matrix avoiding X 1,...,X 4. In this ordering, row θ row (i 1 ) necessarily lies between rows θ row (i 2 ) and θ row (i 3 ) or, else, the submatrix X 4 occurs. Suppose that θ row (i 2 ) <θ row (i 3 ) and θ col (j 1 ) <θ col (j 2 ); then X 3 occurs, a contradiction. The remaining cases are similar. Note that a matrix which avoids X 4 has the consecutive ones property in columns. Gusfield [12, Theorem 3] has proven that a matrix which has an undirected perfect phylogeny can be reordered so as to satisfy this property. In fact, for explainable binary matrices, the reordering used by Gusfield s proof essentially generates a canonical matrix. Note also that Σ-free graphs are bipartite convex as they avoid X 1, X 2, and X 3 (see, e.g., [1]). 4. Algorithms for solving IDP. We now return to the problem of completing an incomplete binary matrix. Let A be the input matrix, and define G(A) = (S, C, E A 1 ). For a nonempty subset S S, we say that a character is S -semiuniversal in A if its 0-set does not intersect S. The following lemmas motivate a divide and conquer approach to IDP, which is the basis of our algorithms for solving it. Lemma 9. Let A be an incomplete matrix with a Σ-free completion B. Let c be S-semiuniversal in A. Let B be the matrix obtained from B by setting all entries in the column of c to 1. Then B is also a Σ-free completion of A. Proof. Suppose to the contrary that {s 1,c 1,s 2,c 2,s 3 } induce a Σ subgraph in G(B ). Since G(B) is Σ-free, if follows that at least one of the Σ edges was added to B. But then one of c 1 and c 2 is c, a contradiction. Lemma 10. Let A be an incomplete matrix with a Σ-free completion B. Let (K 1,...,K r ) be a partition of S C such that each K i is a union of one or more connected components of G(A). Let B be the matrix obtained from B by setting all entries between vertices of K i and K j to 0, for all i j. Then B is also a Σ-free completion of A. Proof. Suppose to the contrary that {s 1,c 1,s 2,c 2,s 3 } induce a Σ subgraph in G(B ). Then one of the nonedges (s 1,c 2 )or(c 1,s 3 ) contains one vertex from K i and the other from K j, for i j. It follows that there is a path in G(B ) between a vertex of K i and a vertex of K j, a contradiction.
9 598 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN Alg A(A =((S, C),E 0,E?,E 1 )): 1. If S > 1 then do: (a) Remove all S-semiuniversal characters and all null characters from G(A). (b) If the resulting graph G is connected, then output False and halt. (c) Otherwise, let K 1,...,K r be the connected components of G, and let A 1,...,A r be the corresponding submatrices of A. (d) For i =1,...,r do: Alg A(A i ). 2. Output S. Fig. 6. Algorithm A for solving IDP. We now describe two efficient Õ(nm)-time algorithms for solving IDP Algorithm A. Algorithm A is described in Figure 6. The algorithm outputs the set of nonempty clades of a tree explaining A, or outputs False if no such tree exists. The algorithm is recursive and is initially called with Alg A(A). Theorem 11. Algorithm A correctly solves IDP. Proof. Suppose that the algorithm outputs False. Then there exists a recursive call Alg A(A ) in which the graph G =(S,C,E ), obtained in Step 1b (see Figure 6), was found to be connected. Suppose to the contrary that A has a phylogenetic tree. Then by Corollary 4, there exists some edge set F, which solves A. The graph G =(S,C,F ) is connected and by Theorem 3, it is also Σ-free. Therefore, by Proposition 6 there exists an S -universal character in G. That character must be S -semiuniversal in A. By Algorithm A this vertex should have been removed at Step 1a, a contradiction. To prove the other direction, we will show that if the algorithm outputs a collection T = {S 1,...,S l } of sets, then T = T { } is a tree which explains A. We first prove that the collection T of sets is pairwise compatible, implying by Observation 1 that T is a tree. Associate with each S i the recursive call Alg A(A i ) at which it was output. Observe that each such call makes recursive calls associated with disjoint subsets of S i. By induction, it follows that S i S j if and only if the recursive call associated with S i is nested within the one associated with S j. Otherwise, S i S j =. Hence, S 1,...,S l are pairwise compatible and, thus, T is a tree. It remains to show that T is a phylogenetic tree for A. Associate each null character with the empty clade. Each other character ĉ is removed at Step 1a only once in the course of the algorithm, during some recursive call Alg A(Â). Associate ĉ with the clade Ŝ which was output at that recursive call. Observe that each nontrivial clade Ŝ T is associated with at least one character. Finally, define a binary matrix B n m with b sc = 1 if and only if s belongs to the clade S c associated with c. Since a sc 1 for all s S c and a sc 0 for all s S c, B is a completion of A. The claim follows. Let h min{m, n} be the height of the reconstructed tree. Each recursive call increases the height of the output tree by at most one. The work at each level of the tree requires (1) Finding semiuniversal vertices and (2) finding connected components in disjoint graphs whose total number of edges is at most mn. Hence, the total work is O(mn) per level, and a naive implementation requires O(hmn) time. We give a faster implementation below.
10 INCOMPLETE DIRECTED PERFECT PHYLOGENY 599 Theorem 12. Algorithm A has a deterministic implementation which takes O(nm+ E 1 log 2 (n+m)) time, and a randomized implementation which takes O(nm+ E 1 log(l 2 / E 1 )+l(log l) 3 log log l) expected time, where l = n + m. Proof. For the complexity proof we give an alternative, nonrecursive implementation of Algorithm A, shown in Figure 7. This iterative version mimics the recursive one, but traverses the tree of recursive calls in a breadth first manner, rather than a depth first manner. Consequently, the implementation deals with a single graph, rather than a different graph per each recursive call. The reduction in complexity is primarily due to the use of an efficient dynamic data structure for graph connectivity. The data structure maintains the connected components of the graph while edge deletions occur. We now analyze the running time of this implementation. Step 1 takes O(nm) time. Each iteration of the while loop (Step 2) splits the (potential) clades added in the previous one. Thus, Algorithm A performs one iteration of this type per each level of the tree returned, and at most h iterations. Step 2b requires explicitly computing the connected components of G. Both data structures that we use for storing the connected components of G (see below) maintain a spanning tree for each connected component of G, and allow computing the connected components in O(n + m) time per iteration, or O(h(m + n)) = O(nm) time in total. The loop of Step 2c is performed at most min{2n 1,m} times altogether, as in each (successful) iteration at least one character is removed from G (Step 2cvii), and at least one clade is added to T. Thus, Step 2ci takes O(min{n, m}) time altogether, and Step 2cii takes O(nm) time in total. Step 2ciii takes O(nm) time in total, as it considers each species-character pair only once throughout the execution of the algorithm. In order to analyze the complexity of Step 2civ, observe that the following invariants hold in this step for each character c C(K i ): d? c = {(s, c) E? s S(K i )}, as guaranteed by Step 2ciii. d 1 c = {(s, c) E 1 s S(K i )} = {(s, c) E 1 s S}, as initialized in Step 1b, since species are never removed, and each species adjacent to c must be in its connected component until c is removed. Given d 1 c,d? c and S(K i ), one can check in O(1) time whether c is S(K i )-semiuniversal, and thus Step 2civ takes O( C(K i ) ) time, or O(hm) time in total. Since each set added to T in Step 2cvi corresponds to at least one character, and each character is associated with exactly one such set, updating T requires O(nm) time in total. This also implies an O(nm) bound on the size of the output produced in Step 3. It remains to discuss the cost of the dynamic data structure, which is charged for Step 2cvii. Using the dynamic algorithm of [15], the connected components of G can be maintained during E 1 edge deletions at a total cost of O( E 1 log 2 (n + m)) time spent in Step 2cvii. Alternatively, using the Las Vegas type randomized algorithm of [23] for decremental dynamic connectivity, the edge deletions can be supported in O( E 1 log(l 2 / E 1 )+l(log l) 3 log log l) expected time. The complexity follows Algorithm B. We now describe another deterministic algorithm for IDP, which is faster than Algorithm A whenever E 1 = ω((n + m) 2 / log(n + m)). Algorithm B uses the dynamic-connectivity data structure of [14], which supports deletion of batches of edges from a graph, while detecting after each batch one of the new connected components in the resulting graph (if new components were formed).
11 600 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN Alg A fast(a =((S, C),E 0,E?,E 1 )): 1. Initialize: (a) Set t 0, K 0 {S C}, G G(A), T triv(s). (b) For each character c, and i {1,?} do: Set d i c {s S (s, c) E i }. (c) Remove all S-semiuniversal and all null characters from G. (d) Initialize a data structure for maintaining the connected components of G. 2. While E(G) do: (a) Increment t. (b) Explicitly compute the set K t of connected components K 1,...,K r of G. (c) For each connected component K i K t such that E(K i ) 1 do: i. Pick any character c C(K i ). ii. Compute S = S(K ) \ S(K i ), where K is the component in K t 1 which contains c. iii. For each species-character pair (s, c) S C(K i ) do: If (s, c) E? then decrement d? c. iv. Compute the set U of all characters in K i which are S(K i )- semiuniversal in A. v. If U =, then output False and halt. vi. Set T T {S(K i )}. vii. Remove U from G and update the data structure of connected components accordingly. 3. Output T. Fig. 7. An iterative presentation of Algorithm A. Algorithm B is described in Figure 8. For an instance A it outputs the nonempty clades of a tree explaining A (except possibly the root clade, if it has no matching character), or False if no such tree exists. It is initially called with Alg B(A). Theorem 13. Algorithm B correctly solves IDP in O((n + m) 2 log(n + m)) deterministic time. Proof. Correctness. We prove correctness by induction on the problem size. If G is connected (at Step 2c), then by Proposition 6 A has no phylogenetic tree, and indeed the algorithm outputs False. Otherwise, let A 1 and A 2 be the subinstances induced on K and K = V (G ) \ K, respectively, as detected in Step 2b. If A has a phylogenetic tree, then by Corollary 4 so do A 1 and A 2. On the other hand, let T 1, T 2 be phylogenetic trees for A 1, A 2, respectively. Note that by definition, T 2 must contain the trivial clade S(K ), which is not necessarily a clade in a phylogenetic tree for A (if K has no semiuniversal character). To remedy that, define T 2 = T 2 if the algorithm outputs S(K ), and T 2 = T 2 \{S(K )} otherwise. Then T 1 T 2 {S} is a phylogenetic tree for A. Complexity. The data structure of [14] dynamically maintains a graph H =(V,E) through batches of edge deletions, with each batch followed by a query for a newly created connected component in the resulting graph. If we denote by b 0 the number of batches which do not result in a new component, then as shown in [14], the total cost of answering the queries and performing the batch deletions, if eventually all edges are deleted, is O( V 2 log V + b 0 min{ V 2, E log V }).
12 INCOMPLETE DIRECTED PERFECT PHYLOGENY 601 Alg B(A =((S, C),E 0,E?,E 1 )): 1. If S =1orG(A) has an S-semiuniversal vertex, then output S. 2. If S > 1, then do: (a) Remove all S-semiuniversal characters and all null characters from G(A). (b) If the resulting graph G contains a new connected component K, then do: i. Let A 1, A 2 be the submatrices of A induced on V (K) and V (G )\ V (K), respectively. ii. For i =1, 2 do: Alg B(A i ). (c) Else output False and halt. Fig. 8. Algorithm B for solving IDP. Characters 1? ?? Species? 1 1??? 1 1? 0? 1 A Characters Species B Fig. 9. A counterexample to the greedy approach. A: The input matrix. B: A solution. We use this data structure to maintain G(A) during all the recursive calls. As b 0 = 1 (since in case no new component is formed the algorithm outputs False and halts) and V = n+m, the total cost is O((m+n) 2 log(n+m)) time. This expression dominates the complexity, as finding the semiuniversal vertices at each recursive call costs in total only O(nm) time (see proof of Theorem 12). We remark that an Ω(nm)-time lower bound for (undirected) binary perfect phylogeny was proved by Gusfield [12]. A closer look at Gusfield s proof reveals that it applies, as is, also to the directed case. As IDP generalizes directed binary perfect phylogeny, any algorithm for this problem would require Ω(nm) time Greedy approach fails. We end the section by showing that a simple greedy approach to IDP fails. Let A be an incomplete matrix. We say that a sc =? is forced if there exists an assignment x {0, 1} such that completing a sc to x results in an induced Σ in the graph (S, C, E1 A E? A ) corresponding to the completed matrix A. A is called forced if it has some forced?-entry. A naive greedy algorithm for IDP is as follows: At each step complete one?- entry in the matrix. If there are no forced entries, choose any?-entry and complete it arbitrarily. Otherwise, try to complete a forced entry. If such completion is not possible (an induced Σ is formed), report False. Figure 9(A) shows an explainable instance with no forced entries. Setting the bottom-left?-entry to 0 results in an instance which cannot be explained. A solution matrix is shown in Figure 9(B). 5. Determining the generality of the solution. A yes instance of IDP may have several distinct phylogenetic trees as solutions. These trees may be related in the following way: We say that a tree T generalizes a tree T, and write T T,
13 602 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN Characters c 1 c 2 Species s 1 s 2 T T 1 T 2 c 1 c 2 c 1 s c 3 1 c 2 c 2 s 1 s 2 s 3 s 1 s 2 s 3 s 1 s2 s 3 c 1 s 1 s 2 s 3 c 1 c 2 c 1 c 2 c 2 s 4 s 5 s 1 s 2 s 3 s 4 s 5 s 1 s 2 s 3 s 4 s 5 Fig. 10. Top left: An IDP instance which has a general solution. Dashed lines denote E? -edges, while solid lines denote E 1 -edges. Top-right: T, T 1, and T 2 are the possible solutions. T generalizes T 1 and T 2 (which are obtained by splitting the root node of T ), and is the general solution. Bottom left: An IDP instance which has no general solution. Bottom middle and bottom right: Two possible solutions. The only tree which generalizes both solutions is the tree composed of the trivial clades only, which is not a solution. if every clade of T is a clade of T ; i.e., the evolutionary scenario expressed by T includes all the details of the scenario expressed by T, and possibly more. Therefore, T represents a more specific scenario, and T represents a more general one. We say that a tree T is the general solution of an instance A, ift explains A and generalizes every other tree which explains A. Figure 10 demonstrates the definitions and also gives an example of an instance that has no general solution. We give in this section a characterization of IDP instances that admit a general solution. We prove that whenever a general solution exists, Algorithm A finds it. We also provide an algorithm to determine whether the solution tree T returned by Algorithm A is general. The complexity of the latter algorithm is shown to be O(mn + E 1 d), where d is the maximum out-degree in T. This notation is used in what follows. Let A be an incomplete matrix and let Ŝ S. We denote by W A (Ŝ) the set of Ŝ-semiuniversal characters in A. Note that if A is binary, then W A (Ŝ) is its set of Ŝ-universal characters. We now define the operator on incomplete matrices: We denote by à the submatrix A S,C\W A (S) of A. In particular, G(Ã) is the graph produced from G(A) by removing its set of S- semiuniversal characters. A species set S S is said to be connected in a graph G, ifs is contained in some connected component of G. Lemma 14. Let T be the general solution for an instance A of IDP. Let S = L(x) be a clade of T, corresponding to some node x. Let T be the subtree of T rooted at x, and let A be the instance induced on S C. Then T is the general solution for A. Proof. By Observation 5, T explains A. Suppose that T also explains A and T T. Then ˆT =(T\T ) T explains A, and T ˆT, a contradiction. A nonempty clade of a tree is called maximal if the only clade that properly contains it is S. Lemma 15. Let T be a phylogenetic tree for a binary matrix B. A nonempty clade S of T is maximal if and only if S is the species set of some connected component of G( B). Proof. Suppose that S is a maximal clade of T. We first claim that S is contained in some connected component K of G( B). If S = 1 this trivially holds. If S > 1,
14 INCOMPLETE DIRECTED PERFECT PHYLOGENY 603 let c be a character associated with S. c is adjacent to all vertices in S and to no other vertex. Hence, c is not S-universal, implying that all the edges {(c, s) :s S } are present in G( B). This proves the claim. It remains to show that S(K) =S. Suppose S(K) S. In particular, S(K) > 1. By Proposition 6, there exists a character c in G( B) whose 1-set is S(K). Hence, S(K) must be a clade of T which is associated with c, contradicting the maximality of S. To prove the converse, let S be the species set of some connected component K of G( B). We first claim that S is a clade. If S =1,S is a trivial clade. Otherwise, by Proposition 6 there exists an S -universal character c in G( B). Since K is a connected component, c has no neighbors in S \ S. Hence, S must be a clade in T. Suppose to the contrary that S is not maximal; then it is properly contained in a maximal clade S, which by the previous direction is the species set of K, a contradiction. Theorem 16. Algorithm A produces the general solution for every IDP instance that has one. Proof. Let A be an instance of IDP for which there exists a general solution T. Let T alg be the solution tree produced by Algorithm A. By definition T T alg. Suppose to the contrary that T T alg. Let S be the largest clade reported by Algorithm A, which is not a clade of T (S must be nontrivial), and let S be the smallest clade in T alg which properly contains S. Let A be the instance induced on S C. By Observation 5, A is explained by the corresponding subtrees T alg of T alg and T of T. By Lemma 14, T is the general solution of A. Due to the recursive nature of Algorithm A, it produces T alg when invoked with input A. Thus, without loss of generality, one can assume that S = S and S is a maximal clade of T alg. Suppose that T explains A via a completion B, and let G = G(B ). Since S is a maximal clade, it is reported during a second level call of Alg A( ) (the call at the first level reports the trivial clade S). Hence, it must be the species set of some connected component K in G(Ã). Since every S-universal character in G is S-semiuniversal in A, S is contained in some connected component K of G( B ). Denote S S(K ). By Lemma 15, S is a maximal clade of T. Since S T, we have S S, and therefore, S S. But T T alg, implying that S is also a nontrivial clade of T alg, in contradiction to the maximality of S. We now characterize IDP instances for which a general solution exists. Let A be a yes instance of IDP. Consider a recursive call Alg A(A ) nested within Alg A(A), where A = A C,S. Let K 1,...,K r be the connected components of G(Ã ), computed in Step 1c. Observe that S(K 1 ),...,S(K r ) are clades to be reported by recursive calls launched during Alg A(A ). A set U of characters is said to be (K i,k j )-critical if characters in U are both S(K i )-semiuniversal and S(K j )-semiuniversal in A, and removing U from G(Ã ) disconnects S(K i ). Note that by definition of U, U W A (S(K i )), and a sc =? for all c U, s S(K j ). A clade S(K i ) is called optional (with respect to A )ifr 3 and there exists a (K i,k j )-critical set for some index j i. IfS(K i ) is not optional we say it is mandatory. In the example of Figure 10 (bottom), let K 1 = {s 1,s 2,c 1 }, K 2 = {s 3 }, and K 3 = {s 4,s 5,c 2 }. The set U = {c 1 } is (K 1,K 2 )- critical, so S(K 1 )={s 1,s 2 } is optional. In contrast, in Figure 10 (top) no clade is optional. Theorem 17. The tree produced by Algorithm A is the general solution if and only if all its clades are mandatory. Proof. Suppose that T alg is the general solution of an instance A. Suppose to the contrary that it contains an optional clade. Without loss of generality, assume it is maximal; i.e., during the recursive call Alg A(A), G = G(Ã) has r 3 connected
15 604 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN components, K 1,...,K r, and there exists a (K i,k j )-critical set U (for some 1 i j r). Let A i, A j and A ij be the subinstances induced on K i,k j and K i K j, respectively. Consider the tree T which is produced by a small modification to the execution of Alg A(A): Instead of recursively invoking Alg A(A i ) and Alg A(A j ), call Alg A(A ij ). Then T is a phylogenetic tree which explains A and includes the clade S(K i K j ). Since removing U from G(Ã) disconnects S(K i), S(K i ) 2so S(K i ) is nontrivial. Moreover, S(K i ) is not a clade of T for the same reason. Hence, T does not contain all clades of T alg, in contradiction to the generality of T alg. Suppose that T alg is not the general solution of an instance A; i.e., there exists a solution T of A such that T alg T. We shall prove the existence of an optional clade in T alg. (The reader is referred to the example in Figure 13 for notation and intuition. The example follows the steps of the proof, leading to the identification of an optional clade.) Let B be a completion of A which is explained by T, and denote G = G(B ). Let S T alg \T be the largest clade reported by Algorithm A which is not a clade of T. Without loss of generality (as argued in the proof of Theorem 16), S is a maximal clade of T alg, and we let S = S(K 1 ), where K 1,...,K r are the connected components of G(Ã). Observe that a binary matrix has at most one phylogenetic tree. Thus, an application of Algorithm A to B necessarily outputs T. Consider such an application, and let {Si }t i=1 be the nested set of reported clades in T which contain S : S = S1 St S (see Figure 11). For each i =1,...,t, let Bi be the instance invoked in the recursive call which reports Si, and let H i be the graph G( B i ), computed in Step 1a of that recursive call. Let Ci be the set of characters in Hi. Equivalently, Ci is the set of characters in Bi whose 1-set is nonempty and is properly contained in Si. Furthermore, define H i to be the subgraph of G(A) induced on Si C i. Observe that H i is the subgraph of G induced on the same vertex set. Since G is a supergraph of G(A), each Hi is a supergraph of H i. Claim 18. S is disconnected in Ht, and therefore also in H t. Proof. Suppose to the contrary that S is contained in some connected component K of Ht. K is thus computed during the tth recursive call (with argument Bt ), and S(K ) is reported as a clade in T by a nested recursive call. Therefore, St S(K ) S, where the first proper containment follows from the fact that Ht is disconnected, and the second from the assumption that S is not a clade of T. Hence, we arrive at a contradiction to the minimality of St. We now return to the proof of Theorem 17. Recall that S is connected in H 1 = G(Ã). Thus, the previous claim implies that t>1. Let K p be a connected component of G(Ã) such that S(K p) S\S2 (see Figure 11). Let l be the minimal index such that there exists some connected component K i of G(Ã) for which S(K i) is disconnected in H l. l is properly defined as S(K 1 ) = S is disconnected in H t. l > 1, since otherwise some K i is disconnected in H 1 and, therefore, also in its subgraph G(Ã), in contradiction to the definition of K 1,...,K r. By minimality of l, Sl S(K i ). Also, Sl St S = S(K 1 ), so Sl S(K i ). We now claim that there exists some connected component K j of G(Ã), j i, such that S(K j ) Sl. Indeed, if i 1, then j =1. Ifi = 1, then l = t (by an argument similar to that in the proof of Claim 18), and since Sl \ S is nonempty, it intersects S(K j ) for some j i. By minimality of l, S(K j ) is properly contained in Sl \ S. Define U W G (Sl ). We now prove that U isa(k i,k j )-critical set. By definition all characters in U are Sl -universal in G, and are thus both K i -semiuniversal
16 INCOMPLETE DIRECTED PERFECT PHYLOGENY 605 S S t S l S 2 S = S 1 S(K p ) Fig. 11. The clades S = S 1 S 2 S t S. S(K i ) S(K j ) S l S(K p ) S U Fig. 12. S(K i ), S(K j ), and S(K p). Note that removing U disconnects S(K i ). and K j -semiuniversal in A. S(K i ) is disconnected in H l = G(A C ). Since K l,s i is a l connected component of G(Ã), S(K i) is disconnected in G(A C l,s ), implying that U isa(k i,k j )-critical set. Also, K i,k j, and K p are distinct, implying that r 3 (see Figure 12). In conclusion, U demonstrates that S(K i ) is optional. The characterization of Theorem 17 leads to an efficient algorithm for determining whether a solution T alg produced by Algorithm A is general. Theorem 19. There is an O(nm + E 1 d)-time algorithm to determine if a given solution T alg is general, where d is the maximum out-degree in T alg. Proof. The algorithm simply traverses T alg bottom-up, searching for optional clades. For each internal node x visited, whose children are y 1,...,y d(x), the algorithm checks whether any of the clades L(y 1 ),...,L(y d(x) ) is optional. If an optional clade is found the algorithm outputs False. Correctness follows from Theorem 17. We show how to efficiently check whether a clade L(y i ) is optional. If d(x) =2, or y i is a leaf, then certainly L(y i ) is mandatory. Otherwise, let U i be the set of characters whose origin (in T alg )isy i. Let Uj i denote the set of characters in U i which are L(y j )-semiuniversal, for j i. The computation of Uj i for all i and j takes O(nm) time in total, since for each character c and species s we check at most once whether (s, c) E? A, for an input instance A. It remains to show how to efficiently check whether for some j, Uj i disconnects L(y i ) in the appropriate subgraph encountered during the execution of Algorithm A. To this end, we define an auxiliary bipartite graph H i whose set of vertices is W i U i,
17 606 I. PE ER, T. PUPKO, R. SHAMIR, AND R. SHARAN Characters c 1 Species s 1 s 2 T alg K p K j K i s 3 c 1 c 2 c 3 c 4 c 2 c 3 s 4 s 5 s 6 s 1 s 2 s 3 T s 4 s 5 s 6 s 7 s 8 S Characters Species s 3 c 4 s 7 c 2 s 4 U s 8 c 1 c 2 c 4 c 3 s 5 c 3 s 6 s 1 s 2 s 3 s 4 s 5 s 6 s 7 s 8 s 7 S = S 1 S 2 S 3 s 8 Fig. 13. An example demonstrating the proof of the if part of Theorem 17, using the notation in the proof. Left: A graphical representation of an input instance A. Dashed lines denote E? -edges, while solid lines denote E 1 -edges. Top right: The tree T alg produced by Algorithm A. Bottom middle: A tree T corresponding to a completion B that uses all the edges in E?. Bottom right: The graphs H 2 (solid edges) and H 2 (solid and dashed edges). T alg T, and S = {s 5,s 6 }. There are t =3 clades of T which contain S : S 1 = {s 1,...,s 8 }, S 2 = {s 3,...,s 8 }, and S 3 = {s 5,s 6,s 7 }. The component K p = {c 1,s 1,s 2 } has its species in S \ S 2. Since W A(S) =W B (S) =, H 1 = G(A). Since W B (S 2 )={c 4}, the species set of the connected component K i = {s 7,s 8,c 4 } is disconnected in H 2, implying that l =2. For a choice of K j = {s 3,s 4,c 2 }, the set U = {c 4 } is (K i,k j )-critical, demonstrating that S is optional. where W i = {w 1,...,w d(yi)} is the set of children of y i in T alg. We include the edge (w r,c p )inh i, for w r W i,c p U i,if(c p,s) E1 A for some species s L(w r ). We construct for each j i a subgraph Hj i of Hi induced on W i (U i \ Uj i ). All we need to report is whether Hj i is connected. For each i we construct H i by considering all E1 A edges connecting characters in U i to species in L(y i ). This takes O( E1 A ) time in total. There are d(y i ) subgraphs Hj i for every y i. Hence, computing Hj i for all j and determining whether each Hi j is connected take O ( E(H i ) d(y i ) ) time. Since i E(Hi ) E1 A, the total time complexity is O(mn + i E(Hi ) d(y i )) = O(mn + E1 A max v Talg d(v)). Acknowledgments. We thank Dan Graur for drawing our attention to this phylogenetic problem, and for helpful discussions. We thank Nati Linial for insightful comments. We thank Joe Felsenstein, Dan Gusfield, Haim Kaplan, Mike Steel, and an anonymous CPM 00 referee for referring us to helpful literature.
Classical Complexity and Fixed-Parameter Tractability of Simultaneous Consecutive Ones Submatrix & Editing Problems
 Classical Complexity and Fixed-Parameter Tractability of Simultaneous Consecutive Ones Submatrix & Editing Problems Rani M. R, Mohith Jagalmohanan, R. Subashini Binary matrices having simultaneous consecutive
Classical Complexity and Fixed-Parameter Tractability of Simultaneous Consecutive Ones Submatrix & Editing Problems Rani M. R, Mohith Jagalmohanan, R. Subashini Binary matrices having simultaneous consecutive
Phylogenetic Networks, Trees, and Clusters
 Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Phylogenetic Networks, Trees, and Clusters Luay Nakhleh 1 and Li-San Wang 2 1 Department of Computer Science Rice University Houston, TX 77005, USA nakhleh@cs.rice.edu 2 Department of Biology University
Strongly chordal and chordal bipartite graphs are sandwich monotone
 Strongly chordal and chordal bipartite graphs are sandwich monotone Pinar Heggernes Federico Mancini Charis Papadopoulos R. Sritharan Abstract A graph class is sandwich monotone if, for every pair of its
Strongly chordal and chordal bipartite graphs are sandwich monotone Pinar Heggernes Federico Mancini Charis Papadopoulos R. Sritharan Abstract A graph class is sandwich monotone if, for every pair of its
Linear-Time Algorithms for Finding Tucker Submatrices and Lekkerkerker-Boland Subgraphs
 Linear-Time Algorithms for Finding Tucker Submatrices and Lekkerkerker-Boland Subgraphs Nathan Lindzey, Ross M. McConnell Colorado State University, Fort Collins CO 80521, USA Abstract. Tucker characterized
Linear-Time Algorithms for Finding Tucker Submatrices and Lekkerkerker-Boland Subgraphs Nathan Lindzey, Ross M. McConnell Colorado State University, Fort Collins CO 80521, USA Abstract. Tucker characterized
Dominating Set Counting in Graph Classes
 Dominating Set Counting in Graph Classes Shuji Kijima 1, Yoshio Okamoto 2, and Takeaki Uno 3 1 Graduate School of Information Science and Electrical Engineering, Kyushu University, Japan kijima@inf.kyushu-u.ac.jp
Dominating Set Counting in Graph Classes Shuji Kijima 1, Yoshio Okamoto 2, and Takeaki Uno 3 1 Graduate School of Information Science and Electrical Engineering, Kyushu University, Japan kijima@inf.kyushu-u.ac.jp
THE THREE-STATE PERFECT PHYLOGENY PROBLEM REDUCES TO 2-SAT
 COMMUNICATIONS IN INFORMATION AND SYSTEMS c 2009 International Press Vol. 9, No. 4, pp. 295-302, 2009 001 THE THREE-STATE PERFECT PHYLOGENY PROBLEM REDUCES TO 2-SAT DAN GUSFIELD AND YUFENG WU Abstract.
COMMUNICATIONS IN INFORMATION AND SYSTEMS c 2009 International Press Vol. 9, No. 4, pp. 295-302, 2009 001 THE THREE-STATE PERFECT PHYLOGENY PROBLEM REDUCES TO 2-SAT DAN GUSFIELD AND YUFENG WU Abstract.
Cluster Graph Modification Problems
 Cluster Graph Modification Problems Ron Shamir Roded Sharan Dekel Tsur December 2002 Abstract In a clustering problem one has to partition a set of elements into homogeneous and well-separated subsets.
Cluster Graph Modification Problems Ron Shamir Roded Sharan Dekel Tsur December 2002 Abstract In a clustering problem one has to partition a set of elements into homogeneous and well-separated subsets.
On improving matchings in trees, via bounded-length augmentations 1
 On improving matchings in trees, via bounded-length augmentations 1 Julien Bensmail a, Valentin Garnero a, Nicolas Nisse a a Université Côte d Azur, CNRS, Inria, I3S, France Abstract Due to a classical
On improving matchings in trees, via bounded-length augmentations 1 Julien Bensmail a, Valentin Garnero a, Nicolas Nisse a a Université Côte d Azur, CNRS, Inria, I3S, France Abstract Due to a classical
Generating p-extremal graphs
 Generating p-extremal graphs Derrick Stolee Department of Mathematics Department of Computer Science University of Nebraska Lincoln s-dstolee1@math.unl.edu August 2, 2011 Abstract Let f(n, p be the maximum
Generating p-extremal graphs Derrick Stolee Department of Mathematics Department of Computer Science University of Nebraska Lincoln s-dstolee1@math.unl.edu August 2, 2011 Abstract Let f(n, p be the maximum
Packing and Covering Dense Graphs
 Packing and Covering Dense Graphs Noga Alon Yair Caro Raphael Yuster Abstract Let d be a positive integer. A graph G is called d-divisible if d divides the degree of each vertex of G. G is called nowhere
Packing and Covering Dense Graphs Noga Alon Yair Caro Raphael Yuster Abstract Let d be a positive integer. A graph G is called d-divisible if d divides the degree of each vertex of G. G is called nowhere
On shredders and vertex connectivity augmentation
 On shredders and vertex connectivity augmentation Gilad Liberman The Open University of Israel giladliberman@gmail.com Zeev Nutov The Open University of Israel nutov@openu.ac.il Abstract We consider the
On shredders and vertex connectivity augmentation Gilad Liberman The Open University of Israel giladliberman@gmail.com Zeev Nutov The Open University of Israel nutov@openu.ac.il Abstract We consider the
A Cubic-Vertex Kernel for Flip Consensus Tree
 To appear in Algorithmica A Cubic-Vertex Kernel for Flip Consensus Tree Christian Komusiewicz Johannes Uhlmann Received: date / Accepted: date Abstract Given a bipartite graph G = (V c, V t, E) and a nonnegative
To appear in Algorithmica A Cubic-Vertex Kernel for Flip Consensus Tree Christian Komusiewicz Johannes Uhlmann Received: date / Accepted: date Abstract Given a bipartite graph G = (V c, V t, E) and a nonnegative
On the mean connected induced subgraph order of cographs
 AUSTRALASIAN JOURNAL OF COMBINATORICS Volume 71(1) (018), Pages 161 183 On the mean connected induced subgraph order of cographs Matthew E Kroeker Lucas Mol Ortrud R Oellermann University of Winnipeg Winnipeg,
AUSTRALASIAN JOURNAL OF COMBINATORICS Volume 71(1) (018), Pages 161 183 On the mean connected induced subgraph order of cographs Matthew E Kroeker Lucas Mol Ortrud R Oellermann University of Winnipeg Winnipeg,
Bounds on parameters of minimally non-linear patterns
 Bounds on parameters of minimally non-linear patterns P.A. CrowdMath Department of Mathematics Massachusetts Institute of Technology Massachusetts, U.S.A. crowdmath@artofproblemsolving.com Submitted: Jan
Bounds on parameters of minimally non-linear patterns P.A. CrowdMath Department of Mathematics Massachusetts Institute of Technology Massachusetts, U.S.A. crowdmath@artofproblemsolving.com Submitted: Jan
Coloring Vertices and Edges of a Path by Nonempty Subsets of a Set
 Coloring Vertices and Edges of a Path by Nonempty Subsets of a Set P.N. Balister E. Győri R.H. Schelp April 28, 28 Abstract A graph G is strongly set colorable if V (G) E(G) can be assigned distinct nonempty
Coloring Vertices and Edges of a Path by Nonempty Subsets of a Set P.N. Balister E. Győri R.H. Schelp April 28, 28 Abstract A graph G is strongly set colorable if V (G) E(G) can be assigned distinct nonempty
A Polynomial-Time Algorithm for Pliable Index Coding
 1 A Polynomial-Time Algorithm for Pliable Index Coding Linqi Song and Christina Fragouli arxiv:1610.06845v [cs.it] 9 Aug 017 Abstract In pliable index coding, we consider a server with m messages and n
1 A Polynomial-Time Algorithm for Pliable Index Coding Linqi Song and Christina Fragouli arxiv:1610.06845v [cs.it] 9 Aug 017 Abstract In pliable index coding, we consider a server with m messages and n
1.3 Vertex Degrees. Vertex Degree for Undirected Graphs: Let G be an undirected. Vertex Degree for Digraphs: Let D be a digraph and y V (D).
 1.3. VERTEX DEGREES 11 1.3 Vertex Degrees Vertex Degree for Undirected Graphs: Let G be an undirected graph and x V (G). The degree d G (x) of x in G: the number of edges incident with x, each loop counting
1.3. VERTEX DEGREES 11 1.3 Vertex Degrees Vertex Degree for Undirected Graphs: Let G be an undirected graph and x V (G). The degree d G (x) of x in G: the number of edges incident with x, each loop counting
Efficient Reassembling of Graphs, Part 1: The Linear Case
 Efficient Reassembling of Graphs, Part 1: The Linear Case Assaf Kfoury Boston University Saber Mirzaei Boston University Abstract The reassembling of a simple connected graph G = (V, E) is an abstraction
Efficient Reassembling of Graphs, Part 1: The Linear Case Assaf Kfoury Boston University Saber Mirzaei Boston University Abstract The reassembling of a simple connected graph G = (V, E) is an abstraction
The Algorithmic Aspects of the Regularity Lemma
 The Algorithmic Aspects of the Regularity Lemma N. Alon R. A. Duke H. Lefmann V. Rödl R. Yuster Abstract The Regularity Lemma of Szemerédi is a result that asserts that every graph can be partitioned in
The Algorithmic Aspects of the Regularity Lemma N. Alon R. A. Duke H. Lefmann V. Rödl R. Yuster Abstract The Regularity Lemma of Szemerédi is a result that asserts that every graph can be partitioned in
Haplotyping as Perfect Phylogeny: A direct approach
 Haplotyping as Perfect Phylogeny: A direct approach Vineet Bafna Dan Gusfield Giuseppe Lancia Shibu Yooseph February 7, 2003 Abstract A full Haplotype Map of the human genome will prove extremely valuable
Haplotyping as Perfect Phylogeny: A direct approach Vineet Bafna Dan Gusfield Giuseppe Lancia Shibu Yooseph February 7, 2003 Abstract A full Haplotype Map of the human genome will prove extremely valuable
arxiv: v1 [cs.ds] 2 Oct 2018
![arxiv: v1 [cs.ds] 2 Oct 2018 arxiv: v1 [cs.ds] 2 Oct 2018](/thumbs/87/96752737.jpg) Contracting to a Longest Path in H-Free Graphs Walter Kern 1 and Daniël Paulusma 2 1 Department of Applied Mathematics, University of Twente, The Netherlands w.kern@twente.nl 2 Department of Computer Science,
Contracting to a Longest Path in H-Free Graphs Walter Kern 1 and Daniël Paulusma 2 1 Department of Applied Mathematics, University of Twente, The Netherlands w.kern@twente.nl 2 Department of Computer Science,
k-blocks: a connectivity invariant for graphs
 1 k-blocks: a connectivity invariant for graphs J. Carmesin R. Diestel M. Hamann F. Hundertmark June 17, 2014 Abstract A k-block in a graph G is a maximal set of at least k vertices no two of which can
1 k-blocks: a connectivity invariant for graphs J. Carmesin R. Diestel M. Hamann F. Hundertmark June 17, 2014 Abstract A k-block in a graph G is a maximal set of at least k vertices no two of which can
arxiv: v1 [math.co] 28 Oct 2016
![arxiv: v1 [math.co] 28 Oct 2016 arxiv: v1 [math.co] 28 Oct 2016](/thumbs/92/108819504.jpg) More on foxes arxiv:1610.09093v1 [math.co] 8 Oct 016 Matthias Kriesell Abstract Jens M. Schmidt An edge in a k-connected graph G is called k-contractible if the graph G/e obtained from G by contracting
More on foxes arxiv:1610.09093v1 [math.co] 8 Oct 016 Matthias Kriesell Abstract Jens M. Schmidt An edge in a k-connected graph G is called k-contractible if the graph G/e obtained from G by contracting
The Complexity of Constructing Evolutionary Trees Using Experiments
 The Complexity of Constructing Evolutionary Trees Using Experiments Gerth Stlting Brodal 1,, Rolf Fagerberg 1,, Christian N. S. Pedersen 1,, and Anna Östlin2, 1 BRICS, Department of Computer Science, University
The Complexity of Constructing Evolutionary Trees Using Experiments Gerth Stlting Brodal 1,, Rolf Fagerberg 1,, Christian N. S. Pedersen 1,, and Anna Östlin2, 1 BRICS, Department of Computer Science, University
Partial cubes: structures, characterizations, and constructions
 Partial cubes: structures, characterizations, and constructions Sergei Ovchinnikov San Francisco State University, Mathematics Department, 1600 Holloway Ave., San Francisco, CA 94132 Abstract Partial cubes
Partial cubes: structures, characterizations, and constructions Sergei Ovchinnikov San Francisco State University, Mathematics Department, 1600 Holloway Ave., San Francisco, CA 94132 Abstract Partial cubes
Triangle-free graphs with no six-vertex induced path
 Triangle-free graphs with no six-vertex induced path Maria Chudnovsky 1, Paul Seymour 2, Sophie Spirkl Princeton University, Princeton, NJ 08544 Mingxian Zhong Columbia University, New York, NY 10027 June
Triangle-free graphs with no six-vertex induced path Maria Chudnovsky 1, Paul Seymour 2, Sophie Spirkl Princeton University, Princeton, NJ 08544 Mingxian Zhong Columbia University, New York, NY 10027 June
More on NP and Reductions
 Indian Institute of Information Technology Design and Manufacturing, Kancheepuram Chennai 600 127, India An Autonomous Institute under MHRD, Govt of India http://www.iiitdm.ac.in COM 501 Advanced Data
Indian Institute of Information Technology Design and Manufacturing, Kancheepuram Chennai 600 127, India An Autonomous Institute under MHRD, Govt of India http://www.iiitdm.ac.in COM 501 Advanced Data
CMSC 451: Lecture 7 Greedy Algorithms for Scheduling Tuesday, Sep 19, 2017
 CMSC CMSC : Lecture Greedy Algorithms for Scheduling Tuesday, Sep 9, 0 Reading: Sects.. and. of KT. (Not covered in DPV.) Interval Scheduling: We continue our discussion of greedy algorithms with a number
CMSC CMSC : Lecture Greedy Algorithms for Scheduling Tuesday, Sep 9, 0 Reading: Sects.. and. of KT. (Not covered in DPV.) Interval Scheduling: We continue our discussion of greedy algorithms with a number
Near-domination in graphs
 Near-domination in graphs Bruce Reed Researcher, Projet COATI, INRIA and Laboratoire I3S, CNRS France, and Visiting Researcher, IMPA, Brazil Alex Scott Mathematical Institute, University of Oxford, Oxford
Near-domination in graphs Bruce Reed Researcher, Projet COATI, INRIA and Laboratoire I3S, CNRS France, and Visiting Researcher, IMPA, Brazil Alex Scott Mathematical Institute, University of Oxford, Oxford
Parameterized Algorithms and Kernels for 3-Hitting Set with Parity Constraints
 Parameterized Algorithms and Kernels for 3-Hitting Set with Parity Constraints Vikram Kamat 1 and Neeldhara Misra 2 1 University of Warsaw vkamat@mimuw.edu.pl 2 Indian Institute of Science, Bangalore neeldhara@csa.iisc.ernet.in
Parameterized Algorithms and Kernels for 3-Hitting Set with Parity Constraints Vikram Kamat 1 and Neeldhara Misra 2 1 University of Warsaw vkamat@mimuw.edu.pl 2 Indian Institute of Science, Bangalore neeldhara@csa.iisc.ernet.in
1 Matchings in Non-Bipartite Graphs
 CS 598CSC: Combinatorial Optimization Lecture date: Feb 9, 010 Instructor: Chandra Chekuri Scribe: Matthew Yancey 1 Matchings in Non-Bipartite Graphs We discuss matching in general undirected graphs. Given
CS 598CSC: Combinatorial Optimization Lecture date: Feb 9, 010 Instructor: Chandra Chekuri Scribe: Matthew Yancey 1 Matchings in Non-Bipartite Graphs We discuss matching in general undirected graphs. Given
Bounded Treewidth Graphs A Survey German Russian Winter School St. Petersburg, Russia
 Bounded Treewidth Graphs A Survey German Russian Winter School St. Petersburg, Russia Andreas Krause krausea@cs.tum.edu Technical University of Munich February 12, 2003 This survey gives an introduction
Bounded Treewidth Graphs A Survey German Russian Winter School St. Petersburg, Russia Andreas Krause krausea@cs.tum.edu Technical University of Munich February 12, 2003 This survey gives an introduction
Graph coloring, perfect graphs
 Lecture 5 (05.04.2013) Graph coloring, perfect graphs Scribe: Tomasz Kociumaka Lecturer: Marcin Pilipczuk 1 Introduction to graph coloring Definition 1. Let G be a simple undirected graph and k a positive
Lecture 5 (05.04.2013) Graph coloring, perfect graphs Scribe: Tomasz Kociumaka Lecturer: Marcin Pilipczuk 1 Introduction to graph coloring Definition 1. Let G be a simple undirected graph and k a positive
Tree sets. Reinhard Diestel
 1 Tree sets Reinhard Diestel Abstract We study an abstract notion of tree structure which generalizes treedecompositions of graphs and matroids. Unlike tree-decompositions, which are too closely linked
1 Tree sets Reinhard Diestel Abstract We study an abstract notion of tree structure which generalizes treedecompositions of graphs and matroids. Unlike tree-decompositions, which are too closely linked
Reconstruction of certain phylogenetic networks from their tree-average distances
 Reconstruction of certain phylogenetic networks from their tree-average distances Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu October 10,
Reconstruction of certain phylogenetic networks from their tree-average distances Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu October 10,
Copyright 2013 Springer Science+Business Media New York
 Meeks, K., and Scott, A. (2014) Spanning trees and the complexity of floodfilling games. Theory of Computing Systems, 54 (4). pp. 731-753. ISSN 1432-4350 Copyright 2013 Springer Science+Business Media
Meeks, K., and Scott, A. (2014) Spanning trees and the complexity of floodfilling games. Theory of Computing Systems, 54 (4). pp. 731-753. ISSN 1432-4350 Copyright 2013 Springer Science+Business Media
Chapter 3 Deterministic planning
 Chapter 3 Deterministic planning In this chapter we describe a number of algorithms for solving the historically most important and most basic type of planning problem. Two rather strong simplifying assumptions
Chapter 3 Deterministic planning In this chapter we describe a number of algorithms for solving the historically most important and most basic type of planning problem. Two rather strong simplifying assumptions
The Reduction of Graph Families Closed under Contraction
 The Reduction of Graph Families Closed under Contraction Paul A. Catlin, Department of Mathematics Wayne State University, Detroit MI 48202 November 24, 2004 Abstract Let S be a family of graphs. Suppose
The Reduction of Graph Families Closed under Contraction Paul A. Catlin, Department of Mathematics Wayne State University, Detroit MI 48202 November 24, 2004 Abstract Let S be a family of graphs. Suppose
On the number of cycles in a graph with restricted cycle lengths
 On the number of cycles in a graph with restricted cycle lengths Dániel Gerbner, Balázs Keszegh, Cory Palmer, Balázs Patkós arxiv:1610.03476v1 [math.co] 11 Oct 2016 October 12, 2016 Abstract Let L be a
On the number of cycles in a graph with restricted cycle lengths Dániel Gerbner, Balázs Keszegh, Cory Palmer, Balázs Patkós arxiv:1610.03476v1 [math.co] 11 Oct 2016 October 12, 2016 Abstract Let L be a
Preliminaries. Graphs. E : set of edges (arcs) (Undirected) Graph : (i, j) = (j, i) (edges) V = {1, 2, 3, 4, 5}, E = {(1, 3), (3, 2), (2, 4)}
 Preliminaries Graphs G = (V, E), V : set of vertices E : set of edges (arcs) (Undirected) Graph : (i, j) = (j, i) (edges) 1 2 3 5 4 V = {1, 2, 3, 4, 5}, E = {(1, 3), (3, 2), (2, 4)} 1 Directed Graph (Digraph)
Preliminaries Graphs G = (V, E), V : set of vertices E : set of edges (arcs) (Undirected) Graph : (i, j) = (j, i) (edges) 1 2 3 5 4 V = {1, 2, 3, 4, 5}, E = {(1, 3), (3, 2), (2, 4)} 1 Directed Graph (Digraph)
1 Some loose ends from last time
 Cornell University, Fall 2010 CS 6820: Algorithms Lecture notes: Kruskal s and Borůvka s MST algorithms September 20, 2010 1 Some loose ends from last time 1.1 A lemma concerning greedy algorithms and
Cornell University, Fall 2010 CS 6820: Algorithms Lecture notes: Kruskal s and Borůvka s MST algorithms September 20, 2010 1 Some loose ends from last time 1.1 A lemma concerning greedy algorithms and
Algorithms Exam TIN093 /DIT602
 Algorithms Exam TIN093 /DIT602 Course: Algorithms Course code: TIN 093, TIN 092 (CTH), DIT 602 (GU) Date, time: 21st October 2017, 14:00 18:00 Building: SBM Responsible teacher: Peter Damaschke, Tel. 5405
Algorithms Exam TIN093 /DIT602 Course: Algorithms Course code: TIN 093, TIN 092 (CTH), DIT 602 (GU) Date, time: 21st October 2017, 14:00 18:00 Building: SBM Responsible teacher: Peter Damaschke, Tel. 5405
Isomorphisms between pattern classes
 Journal of Combinatorics olume 0, Number 0, 1 8, 0000 Isomorphisms between pattern classes M. H. Albert, M. D. Atkinson and Anders Claesson Isomorphisms φ : A B between pattern classes are considered.
Journal of Combinatorics olume 0, Number 0, 1 8, 0000 Isomorphisms between pattern classes M. H. Albert, M. D. Atkinson and Anders Claesson Isomorphisms φ : A B between pattern classes are considered.
The Turán number of sparse spanning graphs
 The Turán number of sparse spanning graphs Noga Alon Raphael Yuster Abstract For a graph H, the extremal number ex(n, H) is the maximum number of edges in a graph of order n not containing a subgraph isomorphic
The Turán number of sparse spanning graphs Noga Alon Raphael Yuster Abstract For a graph H, the extremal number ex(n, H) is the maximum number of edges in a graph of order n not containing a subgraph isomorphic
arxiv: v1 [math.co] 13 May 2016
![arxiv: v1 [math.co] 13 May 2016 arxiv: v1 [math.co] 13 May 2016](/thumbs/94/118742815.jpg) GENERALISED RAMSEY NUMBERS FOR TWO SETS OF CYCLES MIKAEL HANSSON arxiv:1605.04301v1 [math.co] 13 May 2016 Abstract. We determine several generalised Ramsey numbers for two sets Γ 1 and Γ 2 of cycles, in
GENERALISED RAMSEY NUMBERS FOR TWO SETS OF CYCLES MIKAEL HANSSON arxiv:1605.04301v1 [math.co] 13 May 2016 Abstract. We determine several generalised Ramsey numbers for two sets Γ 1 and Γ 2 of cycles, in
Properties of normal phylogenetic networks
 Properties of normal phylogenetic networks Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu August 13, 2009 Abstract. A phylogenetic network is
Properties of normal phylogenetic networks Stephen J. Willson Department of Mathematics Iowa State University Ames, IA 50011 USA swillson@iastate.edu August 13, 2009 Abstract. A phylogenetic network is
Minmax Tree Cover in the Euclidean Space
 Journal of Graph Algorithms and Applications http://jgaa.info/ vol. 15, no. 3, pp. 345 371 (2011) Minmax Tree Cover in the Euclidean Space Seigo Karakawa 1 Ehab Morsy 1 Hiroshi Nagamochi 1 1 Department
Journal of Graph Algorithms and Applications http://jgaa.info/ vol. 15, no. 3, pp. 345 371 (2011) Minmax Tree Cover in the Euclidean Space Seigo Karakawa 1 Ehab Morsy 1 Hiroshi Nagamochi 1 1 Department
Graph Theory. Thomas Bloom. February 6, 2015
 Graph Theory Thomas Bloom February 6, 2015 1 Lecture 1 Introduction A graph (for the purposes of these lectures) is a finite set of vertices, some of which are connected by a single edge. Most importantly,
Graph Theory Thomas Bloom February 6, 2015 1 Lecture 1 Introduction A graph (for the purposes of these lectures) is a finite set of vertices, some of which are connected by a single edge. Most importantly,
FINAL EXAM PRACTICE PROBLEMS CMSC 451 (Spring 2016)
 FINAL EXAM PRACTICE PROBLEMS CMSC 451 (Spring 2016) The final exam will be on Thursday, May 12, from 8:00 10:00 am, at our regular class location (CSI 2117). It will be closed-book and closed-notes, except
FINAL EXAM PRACTICE PROBLEMS CMSC 451 (Spring 2016) The final exam will be on Thursday, May 12, from 8:00 10:00 am, at our regular class location (CSI 2117). It will be closed-book and closed-notes, except
k-protected VERTICES IN BINARY SEARCH TREES
 k-protected VERTICES IN BINARY SEARCH TREES MIKLÓS BÓNA Abstract. We show that for every k, the probability that a randomly selected vertex of a random binary search tree on n nodes is at distance k from
k-protected VERTICES IN BINARY SEARCH TREES MIKLÓS BÓNA Abstract. We show that for every k, the probability that a randomly selected vertex of a random binary search tree on n nodes is at distance k from
arxiv: v1 [cs.dm] 26 Apr 2010
![arxiv: v1 [cs.dm] 26 Apr 2010 arxiv: v1 [cs.dm] 26 Apr 2010](/thumbs/73/68325063.jpg) A Simple Polynomial Algorithm for the Longest Path Problem on Cocomparability Graphs George B. Mertzios Derek G. Corneil arxiv:1004.4560v1 [cs.dm] 26 Apr 2010 Abstract Given a graph G, the longest path
A Simple Polynomial Algorithm for the Longest Path Problem on Cocomparability Graphs George B. Mertzios Derek G. Corneil arxiv:1004.4560v1 [cs.dm] 26 Apr 2010 Abstract Given a graph G, the longest path
Perfect matchings in highly cyclically connected regular graphs
 Perfect matchings in highly cyclically connected regular graphs arxiv:1709.08891v1 [math.co] 6 Sep 017 Robert Lukot ka Comenius University, Bratislava lukotka@dcs.fmph.uniba.sk Edita Rollová University
Perfect matchings in highly cyclically connected regular graphs arxiv:1709.08891v1 [math.co] 6 Sep 017 Robert Lukot ka Comenius University, Bratislava lukotka@dcs.fmph.uniba.sk Edita Rollová University
An Algebraic View of the Relation between Largest Common Subtrees and Smallest Common Supertrees
 An Algebraic View of the Relation between Largest Common Subtrees and Smallest Common Supertrees Francesc Rosselló 1, Gabriel Valiente 2 1 Department of Mathematics and Computer Science, Research Institute
An Algebraic View of the Relation between Largest Common Subtrees and Smallest Common Supertrees Francesc Rosselló 1, Gabriel Valiente 2 1 Department of Mathematics and Computer Science, Research Institute
A Separator Theorem for Graphs with an Excluded Minor and its Applications
 A Separator Theorem for Graphs with an Excluded Minor and its Applications Noga Alon IBM Almaden Research Center, San Jose, CA 95120,USA and Sackler Faculty of Exact Sciences, Tel Aviv University, Tel
A Separator Theorem for Graphs with an Excluded Minor and its Applications Noga Alon IBM Almaden Research Center, San Jose, CA 95120,USA and Sackler Faculty of Exact Sciences, Tel Aviv University, Tel
Combinatorial Optimization
 Combinatorial Optimization 2017-2018 1 Maximum matching on bipartite graphs Given a graph G = (V, E), find a maximum cardinal matching. 1.1 Direct algorithms Theorem 1.1 (Petersen, 1891) A matching M is
Combinatorial Optimization 2017-2018 1 Maximum matching on bipartite graphs Given a graph G = (V, E), find a maximum cardinal matching. 1.1 Direct algorithms Theorem 1.1 (Petersen, 1891) A matching M is
Induced Subgraph Isomorphism on proper interval and bipartite permutation graphs
 Induced Subgraph Isomorphism on proper interval and bipartite permutation graphs Pinar Heggernes Pim van t Hof Daniel Meister Yngve Villanger Abstract Given two graphs G and H as input, the Induced Subgraph
Induced Subgraph Isomorphism on proper interval and bipartite permutation graphs Pinar Heggernes Pim van t Hof Daniel Meister Yngve Villanger Abstract Given two graphs G and H as input, the Induced Subgraph
5 Flows and cuts in digraphs
 5 Flows and cuts in digraphs Recall that a digraph or network is a pair G = (V, E) where V is a set and E is a multiset of ordered pairs of elements of V, which we refer to as arcs. Note that two vertices
5 Flows and cuts in digraphs Recall that a digraph or network is a pair G = (V, E) where V is a set and E is a multiset of ordered pairs of elements of V, which we refer to as arcs. Note that two vertices
Coloring Vertices and Edges of a Path by Nonempty Subsets of a Set
 Coloring Vertices and Edges of a Path by Nonempty Subsets of a Set P.N. Balister E. Győri R.H. Schelp November 8, 28 Abstract A graph G is strongly set colorable if V (G) E(G) can be assigned distinct
Coloring Vertices and Edges of a Path by Nonempty Subsets of a Set P.N. Balister E. Győri R.H. Schelp November 8, 28 Abstract A graph G is strongly set colorable if V (G) E(G) can be assigned distinct
The concentration of the chromatic number of random graphs
 The concentration of the chromatic number of random graphs Noga Alon Michael Krivelevich Abstract We prove that for every constant δ > 0 the chromatic number of the random graph G(n, p) with p = n 1/2
The concentration of the chromatic number of random graphs Noga Alon Michael Krivelevich Abstract We prove that for every constant δ > 0 the chromatic number of the random graph G(n, p) with p = n 1/2
arxiv: v1 [cs.dm] 29 Oct 2012
![arxiv: v1 [cs.dm] 29 Oct 2012 arxiv: v1 [cs.dm] 29 Oct 2012](/thumbs/85/91540498.jpg) arxiv:1210.7684v1 [cs.dm] 29 Oct 2012 Square-Root Finding Problem In Graphs, A Complete Dichotomy Theorem. Babak Farzad 1 and Majid Karimi 2 Department of Mathematics Brock University, St. Catharines,
arxiv:1210.7684v1 [cs.dm] 29 Oct 2012 Square-Root Finding Problem In Graphs, A Complete Dichotomy Theorem. Babak Farzad 1 and Majid Karimi 2 Department of Mathematics Brock University, St. Catharines,
Berge Trigraphs. Maria Chudnovsky 1 Princeton University, Princeton NJ March 15, 2004; revised December 2, Research Fellow.
 Berge Trigraphs Maria Chudnovsky 1 Princeton University, Princeton NJ 08544 March 15, 2004; revised December 2, 2005 1 This research was partially conducted during the period the author served as a Clay
Berge Trigraphs Maria Chudnovsky 1 Princeton University, Princeton NJ 08544 March 15, 2004; revised December 2, 2005 1 This research was partially conducted during the period the author served as a Clay
THE MAXIMAL SUBGROUPS AND THE COMPLEXITY OF THE FLOW SEMIGROUP OF FINITE (DI)GRAPHS
 THE MAXIMAL SUBGROUPS AND THE COMPLEXITY OF THE FLOW SEMIGROUP OF FINITE (DI)GRAPHS GÁBOR HORVÁTH, CHRYSTOPHER L. NEHANIV, AND KÁROLY PODOSKI Dedicated to John Rhodes on the occasion of his 80th birthday.
THE MAXIMAL SUBGROUPS AND THE COMPLEXITY OF THE FLOW SEMIGROUP OF FINITE (DI)GRAPHS GÁBOR HORVÁTH, CHRYSTOPHER L. NEHANIV, AND KÁROLY PODOSKI Dedicated to John Rhodes on the occasion of his 80th birthday.
MATHEMATICAL ENGINEERING TECHNICAL REPORTS. Boundary cliques, clique trees and perfect sequences of maximal cliques of a chordal graph
 MATHEMATICAL ENGINEERING TECHNICAL REPORTS Boundary cliques, clique trees and perfect sequences of maximal cliques of a chordal graph Hisayuki HARA and Akimichi TAKEMURA METR 2006 41 July 2006 DEPARTMENT
MATHEMATICAL ENGINEERING TECHNICAL REPORTS Boundary cliques, clique trees and perfect sequences of maximal cliques of a chordal graph Hisayuki HARA and Akimichi TAKEMURA METR 2006 41 July 2006 DEPARTMENT
A Single-Exponential Fixed-Parameter Algorithm for Distance-Hereditary Vertex Deletion
 A Single-Exponential Fixed-Parameter Algorithm for Distance-Hereditary Vertex Deletion Eduard Eiben a, Robert Ganian a, O-joung Kwon b a Algorithms and Complexity Group, TU Wien, Vienna, Austria b Logic
A Single-Exponential Fixed-Parameter Algorithm for Distance-Hereditary Vertex Deletion Eduard Eiben a, Robert Ganian a, O-joung Kwon b a Algorithms and Complexity Group, TU Wien, Vienna, Austria b Logic
Complexity Theory VU , SS The Polynomial Hierarchy. Reinhard Pichler
 Complexity Theory Complexity Theory VU 181.142, SS 2018 6. The Polynomial Hierarchy Reinhard Pichler Institut für Informationssysteme Arbeitsbereich DBAI Technische Universität Wien 15 May, 2018 Reinhard
Complexity Theory Complexity Theory VU 181.142, SS 2018 6. The Polynomial Hierarchy Reinhard Pichler Institut für Informationssysteme Arbeitsbereich DBAI Technische Universität Wien 15 May, 2018 Reinhard
Outline. Complexity Theory EXACT TSP. The Class DP. Definition. Problem EXACT TSP. Complexity of EXACT TSP. Proposition VU 181.
 Complexity Theory Complexity Theory Outline Complexity Theory VU 181.142, SS 2018 6. The Polynomial Hierarchy Reinhard Pichler Institut für Informationssysteme Arbeitsbereich DBAI Technische Universität
Complexity Theory Complexity Theory Outline Complexity Theory VU 181.142, SS 2018 6. The Polynomial Hierarchy Reinhard Pichler Institut für Informationssysteme Arbeitsbereich DBAI Technische Universität
Evolutionary Tree Analysis. Overview
 CSI/BINF 5330 Evolutionary Tree Analysis Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Distance-Based Evolutionary Tree Reconstruction Character-Based
CSI/BINF 5330 Evolutionary Tree Analysis Young-Rae Cho Associate Professor Department of Computer Science Baylor University Overview Backgrounds Distance-Based Evolutionary Tree Reconstruction Character-Based
arxiv: v1 [math.co] 11 Jul 2016
![arxiv: v1 [math.co] 11 Jul 2016 arxiv: v1 [math.co] 11 Jul 2016](/thumbs/93/113731245.jpg) Characterization and recognition of proper tagged probe interval graphs Sourav Chakraborty, Shamik Ghosh, Sanchita Paul and Malay Sen arxiv:1607.02922v1 [math.co] 11 Jul 2016 October 29, 2018 Abstract
Characterization and recognition of proper tagged probe interval graphs Sourav Chakraborty, Shamik Ghosh, Sanchita Paul and Malay Sen arxiv:1607.02922v1 [math.co] 11 Jul 2016 October 29, 2018 Abstract
Approximation algorithms for cycle packing problems
 Approximation algorithms for cycle packing problems Michael Krivelevich Zeev Nutov Raphael Yuster Abstract The cycle packing number ν c (G) of a graph G is the maximum number of pairwise edgedisjoint cycles
Approximation algorithms for cycle packing problems Michael Krivelevich Zeev Nutov Raphael Yuster Abstract The cycle packing number ν c (G) of a graph G is the maximum number of pairwise edgedisjoint cycles
4-coloring P 6 -free graphs with no induced 5-cycles
 4-coloring P 6 -free graphs with no induced 5-cycles Maria Chudnovsky Department of Mathematics, Princeton University 68 Washington Rd, Princeton NJ 08544, USA mchudnov@math.princeton.edu Peter Maceli,
4-coloring P 6 -free graphs with no induced 5-cycles Maria Chudnovsky Department of Mathematics, Princeton University 68 Washington Rd, Princeton NJ 08544, USA mchudnov@math.princeton.edu Peter Maceli,
Computer Science & Engineering 423/823 Design and Analysis of Algorithms
 Computer Science & Engineering 423/823 Design and Analysis of Algorithms Lecture 03 Dynamic Programming (Chapter 15) Stephen Scott and Vinodchandran N. Variyam sscott@cse.unl.edu 1/44 Introduction Dynamic
Computer Science & Engineering 423/823 Design and Analysis of Algorithms Lecture 03 Dynamic Programming (Chapter 15) Stephen Scott and Vinodchandran N. Variyam sscott@cse.unl.edu 1/44 Introduction Dynamic
A Phylogenetic Network Construction due to Constrained Recombination
 A Phylogenetic Network Construction due to Constrained Recombination Mohd. Abdul Hai Zahid Research Scholar Research Supervisors: Dr. R.C. Joshi Dr. Ankush Mittal Department of Electronics and Computer
A Phylogenetic Network Construction due to Constrained Recombination Mohd. Abdul Hai Zahid Research Scholar Research Supervisors: Dr. R.C. Joshi Dr. Ankush Mittal Department of Electronics and Computer
Vertex colorings of graphs without short odd cycles
 Vertex colorings of graphs without short odd cycles Andrzej Dudek and Reshma Ramadurai Department of Mathematical Sciences Carnegie Mellon University Pittsburgh, PA 1513, USA {adudek,rramadur}@andrew.cmu.edu
Vertex colorings of graphs without short odd cycles Andrzej Dudek and Reshma Ramadurai Department of Mathematical Sciences Carnegie Mellon University Pittsburgh, PA 1513, USA {adudek,rramadur}@andrew.cmu.edu
Cleaning Interval Graphs
 Cleaning Interval Graphs Dániel Marx and Ildikó Schlotter Department of Computer Science and Information Theory, Budapest University of Technology and Economics, H-1521 Budapest, Hungary. {dmarx,ildi}@cs.bme.hu
Cleaning Interval Graphs Dániel Marx and Ildikó Schlotter Department of Computer Science and Information Theory, Budapest University of Technology and Economics, H-1521 Budapest, Hungary. {dmarx,ildi}@cs.bme.hu
The chromatic number of ordered graphs with constrained conflict graphs
 AUSTRALASIAN JOURNAL OF COMBINATORICS Volume 69(1 (017, Pages 74 104 The chromatic number of ordered graphs with constrained conflict graphs Maria Axenovich Jonathan Rollin Torsten Ueckerdt Department
AUSTRALASIAN JOURNAL OF COMBINATORICS Volume 69(1 (017, Pages 74 104 The chromatic number of ordered graphs with constrained conflict graphs Maria Axenovich Jonathan Rollin Torsten Ueckerdt Department
Robust Network Codes for Unicast Connections: A Case Study
 Robust Network Codes for Unicast Connections: A Case Study Salim Y. El Rouayheb, Alex Sprintson, and Costas Georghiades Department of Electrical and Computer Engineering Texas A&M University College Station,
Robust Network Codes for Unicast Connections: A Case Study Salim Y. El Rouayheb, Alex Sprintson, and Costas Georghiades Department of Electrical and Computer Engineering Texas A&M University College Station,
Online Interval Coloring and Variants
 Online Interval Coloring and Variants Leah Epstein 1, and Meital Levy 1 Department of Mathematics, University of Haifa, 31905 Haifa, Israel. Email: lea@math.haifa.ac.il School of Computer Science, Tel-Aviv
Online Interval Coloring and Variants Leah Epstein 1, and Meital Levy 1 Department of Mathematics, University of Haifa, 31905 Haifa, Israel. Email: lea@math.haifa.ac.il School of Computer Science, Tel-Aviv
Countable Menger s theorem with finitary matroid constraints on the ingoing edges
 Countable Menger s theorem with finitary matroid constraints on the ingoing edges Attila Joó Alfréd Rényi Institute of Mathematics, MTA-ELTE Egerváry Research Group. Budapest, Hungary jooattila@renyi.hu
Countable Menger s theorem with finitary matroid constraints on the ingoing edges Attila Joó Alfréd Rényi Institute of Mathematics, MTA-ELTE Egerváry Research Group. Budapest, Hungary jooattila@renyi.hu
CSCE 750 Final Exam Answer Key Wednesday December 7, 2005
 CSCE 750 Final Exam Answer Key Wednesday December 7, 2005 Do all problems. Put your answers on blank paper or in a test booklet. There are 00 points total in the exam. You have 80 minutes. Please note
CSCE 750 Final Exam Answer Key Wednesday December 7, 2005 Do all problems. Put your answers on blank paper or in a test booklet. There are 00 points total in the exam. You have 80 minutes. Please note
arxiv:quant-ph/ v1 15 Apr 2005
 Quantum walks on directed graphs Ashley Montanaro arxiv:quant-ph/0504116v1 15 Apr 2005 February 1, 2008 Abstract We consider the definition of quantum walks on directed graphs. Call a directed graph reversible
Quantum walks on directed graphs Ashley Montanaro arxiv:quant-ph/0504116v1 15 Apr 2005 February 1, 2008 Abstract We consider the definition of quantum walks on directed graphs. Call a directed graph reversible
A note on network reliability
 A note on network reliability Noga Alon Institute for Advanced Study, Princeton, NJ 08540 and Department of Mathematics Tel Aviv University, Tel Aviv, Israel Let G = (V, E) be a loopless undirected multigraph,
A note on network reliability Noga Alon Institute for Advanced Study, Princeton, NJ 08540 and Department of Mathematics Tel Aviv University, Tel Aviv, Israel Let G = (V, E) be a loopless undirected multigraph,
Some hard families of parameterised counting problems
 Some hard families of parameterised counting problems Mark Jerrum and Kitty Meeks School of Mathematical Sciences, Queen Mary University of London {m.jerrum,k.meeks}@qmul.ac.uk September 2014 Abstract
Some hard families of parameterised counting problems Mark Jerrum and Kitty Meeks School of Mathematical Sciences, Queen Mary University of London {m.jerrum,k.meeks}@qmul.ac.uk September 2014 Abstract
Rao s degree sequence conjecture
 Rao s degree sequence conjecture Maria Chudnovsky 1 Columbia University, New York, NY 10027 Paul Seymour 2 Princeton University, Princeton, NJ 08544 July 31, 2009; revised December 10, 2013 1 Supported
Rao s degree sequence conjecture Maria Chudnovsky 1 Columbia University, New York, NY 10027 Paul Seymour 2 Princeton University, Princeton, NJ 08544 July 31, 2009; revised December 10, 2013 1 Supported
The Chromatic Number of Ordered Graphs With Constrained Conflict Graphs
 The Chromatic Number of Ordered Graphs With Constrained Conflict Graphs Maria Axenovich and Jonathan Rollin and Torsten Ueckerdt September 3, 016 Abstract An ordered graph G is a graph whose vertex set
The Chromatic Number of Ordered Graphs With Constrained Conflict Graphs Maria Axenovich and Jonathan Rollin and Torsten Ueckerdt September 3, 016 Abstract An ordered graph G is a graph whose vertex set
Corrigendum: The complexity of counting graph homomorphisms
 Corrigendum: The complexity of counting graph homomorphisms Martin Dyer School of Computing University of Leeds Leeds, U.K. LS2 9JT dyer@comp.leeds.ac.uk Catherine Greenhill School of Mathematics The University
Corrigendum: The complexity of counting graph homomorphisms Martin Dyer School of Computing University of Leeds Leeds, U.K. LS2 9JT dyer@comp.leeds.ac.uk Catherine Greenhill School of Mathematics The University
Optimal Tree-decomposition Balancing and Reachability on Low Treewidth Graphs
 Optimal Tree-decomposition Balancing and Reachability on Low Treewidth Graphs Krishnendu Chatterjee Rasmus Ibsen-Jensen Andreas Pavlogiannis IST Austria Abstract. We consider graphs with n nodes together
Optimal Tree-decomposition Balancing and Reachability on Low Treewidth Graphs Krishnendu Chatterjee Rasmus Ibsen-Jensen Andreas Pavlogiannis IST Austria Abstract. We consider graphs with n nodes together
Bulletin of the Iranian Mathematical Society
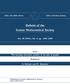 ISSN: 117-6X (Print) ISSN: 1735-8515 (Online) Bulletin of the Iranian Mathematical Society Vol. 4 (14), No. 6, pp. 1491 154. Title: The locating chromatic number of the join of graphs Author(s): A. Behtoei
ISSN: 117-6X (Print) ISSN: 1735-8515 (Online) Bulletin of the Iranian Mathematical Society Vol. 4 (14), No. 6, pp. 1491 154. Title: The locating chromatic number of the join of graphs Author(s): A. Behtoei
Maximising the number of induced cycles in a graph
 Maximising the number of induced cycles in a graph Natasha Morrison Alex Scott April 12, 2017 Abstract We determine the maximum number of induced cycles that can be contained in a graph on n n 0 vertices,
Maximising the number of induced cycles in a graph Natasha Morrison Alex Scott April 12, 2017 Abstract We determine the maximum number of induced cycles that can be contained in a graph on n n 0 vertices,
Contents. Introduction
 Introduction Contents Chapter 1. Network Flows 1 1.1. Graphs 1 1.2. Shortest Paths 8 1.3. Maximum Flow 17 1.4. Min Cost Flows 22 List of frequently used Symbols 31 Bibliography 33 Index 37 iii i Introduction
Introduction Contents Chapter 1. Network Flows 1 1.1. Graphs 1 1.2. Shortest Paths 8 1.3. Maximum Flow 17 1.4. Min Cost Flows 22 List of frequently used Symbols 31 Bibliography 33 Index 37 iii i Introduction
Some Nordhaus-Gaddum-type Results
 Some Nordhaus-Gaddum-type Results Wayne Goddard Department of Mathematics Massachusetts Institute of Technology Cambridge, USA Michael A. Henning Department of Mathematics University of Natal Pietermaritzburg,
Some Nordhaus-Gaddum-type Results Wayne Goddard Department of Mathematics Massachusetts Institute of Technology Cambridge, USA Michael A. Henning Department of Mathematics University of Natal Pietermaritzburg,
Automorphism groups of wreath product digraphs
 Automorphism groups of wreath product digraphs Edward Dobson Department of Mathematics and Statistics Mississippi State University PO Drawer MA Mississippi State, MS 39762 USA dobson@math.msstate.edu Joy
Automorphism groups of wreath product digraphs Edward Dobson Department of Mathematics and Statistics Mississippi State University PO Drawer MA Mississippi State, MS 39762 USA dobson@math.msstate.edu Joy
MATROID PACKING AND COVERING WITH CIRCUITS THROUGH AN ELEMENT
 MATROID PACKING AND COVERING WITH CIRCUITS THROUGH AN ELEMENT MANOEL LEMOS AND JAMES OXLEY Abstract. In 1981, Seymour proved a conjecture of Welsh that, in a connected matroid M, the sum of the maximum
MATROID PACKING AND COVERING WITH CIRCUITS THROUGH AN ELEMENT MANOEL LEMOS AND JAMES OXLEY Abstract. In 1981, Seymour proved a conjecture of Welsh that, in a connected matroid M, the sum of the maximum
Even Cycles in Hypergraphs.
 Even Cycles in Hypergraphs. Alexandr Kostochka Jacques Verstraëte Abstract A cycle in a hypergraph A is an alternating cyclic sequence A 0, v 0, A 1, v 1,..., A k 1, v k 1, A 0 of distinct edges A i and
Even Cycles in Hypergraphs. Alexandr Kostochka Jacques Verstraëte Abstract A cycle in a hypergraph A is an alternating cyclic sequence A 0, v 0, A 1, v 1,..., A k 1, v k 1, A 0 of distinct edges A i and
Paths and cycles in extended and decomposable digraphs
 Paths and cycles in extended and decomposable digraphs Jørgen Bang-Jensen Gregory Gutin Department of Mathematics and Computer Science Odense University, Denmark Abstract We consider digraphs called extended
Paths and cycles in extended and decomposable digraphs Jørgen Bang-Jensen Gregory Gutin Department of Mathematics and Computer Science Odense University, Denmark Abstract We consider digraphs called extended
Out-colourings of Digraphs
 Out-colourings of Digraphs N. Alon J. Bang-Jensen S. Bessy July 13, 2017 Abstract We study vertex colourings of digraphs so that no out-neighbourhood is monochromatic and call such a colouring an out-colouring.
Out-colourings of Digraphs N. Alon J. Bang-Jensen S. Bessy July 13, 2017 Abstract We study vertex colourings of digraphs so that no out-neighbourhood is monochromatic and call such a colouring an out-colouring.
Limitations of Algorithm Power
 Limitations of Algorithm Power Objectives We now move into the third and final major theme for this course. 1. Tools for analyzing algorithms. 2. Design strategies for designing algorithms. 3. Identifying
Limitations of Algorithm Power Objectives We now move into the third and final major theme for this course. 1. Tools for analyzing algorithms. 2. Design strategies for designing algorithms. 3. Identifying
Aditya Bhaskara CS 5968/6968, Lecture 1: Introduction and Review 12 January 2016
 Lecture 1: Introduction and Review We begin with a short introduction to the course, and logistics. We then survey some basics about approximation algorithms and probability. We also introduce some of
Lecture 1: Introduction and Review We begin with a short introduction to the course, and logistics. We then survey some basics about approximation algorithms and probability. We also introduce some of
4 CONNECTED PROJECTIVE-PLANAR GRAPHS ARE HAMILTONIAN. Robin Thomas* Xingxing Yu**
 4 CONNECTED PROJECTIVE-PLANAR GRAPHS ARE HAMILTONIAN Robin Thomas* Xingxing Yu** School of Mathematics Georgia Institute of Technology Atlanta, Georgia 30332, USA May 1991, revised 23 October 1993. Published
4 CONNECTED PROJECTIVE-PLANAR GRAPHS ARE HAMILTONIAN Robin Thomas* Xingxing Yu** School of Mathematics Georgia Institute of Technology Atlanta, Georgia 30332, USA May 1991, revised 23 October 1993. Published
On colorability of graphs with forbidden minors along paths and circuits
 On colorability of graphs with forbidden minors along paths and circuits Elad Horev horevel@cs.bgu.ac.il Department of Computer Science Ben-Gurion University of the Negev Beer-Sheva 84105, Israel Abstract.
On colorability of graphs with forbidden minors along paths and circuits Elad Horev horevel@cs.bgu.ac.il Department of Computer Science Ben-Gurion University of the Negev Beer-Sheva 84105, Israel Abstract.
Partitioning a graph into highly connected subgraphs
 Partitioning a graph into highly connected subgraphs Valentin Borozan 1,5, Michael Ferrara, Shinya Fujita 3 Michitaka Furuya 4, Yannis Manoussakis 5, Narayanan N 6 and Derrick Stolee 7 Abstract Given k
Partitioning a graph into highly connected subgraphs Valentin Borozan 1,5, Michael Ferrara, Shinya Fujita 3 Michitaka Furuya 4, Yannis Manoussakis 5, Narayanan N 6 and Derrick Stolee 7 Abstract Given k
