Supporting information for Demographic history and rare allele sharing among human populations.
|
|
|
- Raymond Higgins
- 5 years ago
- Views:
Transcription
1 Supporting information for Demographic history and rare allele sharing among human populations. Simon Gravel, Brenna M. Henn, Ryan N. Gutenkunst, mit R. Indap, Gabor T. Marth, ndrew G. Clark, The 1 Genomes consortium, Carlos D. Bustamante I. ESTIMTING PRMETER CONFIDENCE INTERVLS VI BOOTSTRP In order to obtain confidence intervals on the inferred demographic parameters, we used a bootstrap approach. Since linkage effectively reduces the number of independent measurements, we re-sampled genomic regions rather than individual SNPs. More specifically, we considered regions with a coding sequence CDS annotation in Gencode [1] version 3b, and merged regions within 5kbp of each other. We were then left with 9328 distinct and well-separated regions. We resampled these with replacement, forming a set of overlapping genomic regions. For each of these bootstrapped sets, we extracted SNPs generated from both the high- and low-coverage data. We obtained inferred values for both the error rates due to low coverage and the resulting inferred demographic parameters. N1819, N18112, N18147, N1867] 5 samples from CHD, and [N1946, N1937, N1931, N19312, N19317, N19321, N19441, N19453, N19456] 9 samples from LWK. III. ERROR CORRECTION The matrix describing the error model can be inverted to give a correction model with Sijk = 1 S ijk;i j k i j k, i j k 1 ijk;i j k = 1 1 ii 2 1 jj 3 1 kk S1 II. SMPLE SELECTION and Short read sequencing results in high site-to-site and sample-to-sample variability in coverage beyond the independent draw, Poisson sampling [Marth et al., in submission]. Since we are interested in obtaining high quality reads from the capture data, we discarded individuals for which the mean over exons of the median coverage per exon was below 15. We also discarded individuals with unusually high discrepancy with validation or HapMap data. s a result, we removed samples [N18524, N18529, N1853, N18531, N18543, N18557, N18599, N18615, N1862, N18627, N18628, N18635, N18642, N18749, N18773] 15 samples from CHB, [N6985, N6994, N1184, N11995, N124, N126, N12156, N12414, N12763, N12815, N12829, N12842, N12872, N12873, N12874, N12889] 16 samples from CEU, [N18948, N18957, N18961, N18964, N18969, N18979, N18981, N18985, N18988, N18989, N18994, N196, N1954, N1962, N1965, N1968, N1977, N199, N19554, N19559, N19562] 21 samples from JPT and [N18489, N18499, N1854, N18516, N18522, N18865, N1887, N18871, N18917, N1912, N19116, N19137, N19141, N19181, N1921, N1927, N1921, N1922] 18 samples from YRI, for a total of 7 samples in the 4 populations sequenced in the low-coverage pilot. For exononly analysis, we also removed, for the same reasons, samples [N2511, N2512, N2518, N2519, N2524, N2527, N2528, N2529, N253, N254, N2587, N2763] 12 samples from TSI [N17966, p 1 ii = 1 1 ɛ p i if i = i ɛ p i 1 ɛ p i if i =, i otherwise. S2 Note p ii 1 = for i =, i, implying that it is not necessary to know the number of observed invariant sites to estimate the corrected SFS for variable sites. The same holds true for, obtaining Sijk for i, j, k,, does not require the knowledge of S,,. In practice, this correction model should be used with caution; in some cases, estimated entries in the corrected SFS result from a difference of two large numbers, and can even be negative. The Poisson random field approximation, used in the maximum likelihood calculation, would not hold for such cases. However, the corrected model is a sensible approach for calculating basic population demographic estimates such as F ST that are not based on likelihood estimates. Comparison of exon data, low-coverage data, and this errorcorrection model are shown on Figures S1, S2, S3. IV. COMPRISON BETWEEN MODEL ND DT FOR MRGINL FREQUENCY SPECTR. We compare in Figure S5 the model predicted and marginal site frequency spectra for the CEU, CHB+JPT, and YRI panels.
2 2 1 8 YRI low coverage pilot exon pilot corrected low coverage pilot 1 8 CHB+JPT low coverage pilot exon pilot corrected low coverage pilot counts in target region 6 4 counts in target region derived allele counts out of derived allele counts out of 8 FIG. S1. Site frequency spectra for the YRI panel for sites with variant calls in the exon pilot. Shown are the exon pilot data, the lowcoverage data, and a corrected SFS using the exponentially decaying error model, for derived allele frequency below 25%. ll spectra are polarized using the March 26 assembly of the chimp genome PanTro2. The corrected SFS captures the bulk of the differences between the exon and low coverage data across the population frequencies. FIG. S3. Site frequency spectra for the CHB and JPT panels for sites with variant calls in the exon pilot. Shown are the exon pilot data, the low-coverage data, and a corrected SFS using the exponentially decaying error model, for derived allele frequency below 25%. ll spectra are polarized using the March 26 assembly of the chimp genome PanTro2. The corrected SFS captures the bulk of the differences between the exon and low coverage data across the population frequencies. counts in target region CEU low coverage pilot exon pilot corrected low coverage pilot derived allele counts out of 8 FIG. S2. Site frequency spectra for the CEU panel for sites with variant calls in the exon pilot. Shown are the exon pilot data, the lowcoverage data, and a corrected SFS using the exponentially decaying error model, for derived allele frequency below 25%. ll spectra are polarized using the March 26 assembly of the chimp genome PanTro2. The corrected SFS captures the bulk of the differences between the exon and low coverage data across the population frequencies. V. JCKKNIFE EXPNSION The Burnham-Overton jacknife was introduced to predict the number of unobserved objects typically, animals based on a finite number of mark-and-recapture field trips. The basic assumption of this model is a postulated relation between the total number of objects V, the number V n of objects observed after n field trips, and n itself : ˆV = V n + p i=1 a p i n i. The Burnham-Overton jackknife estimator has been shown to perform well in a variety of situations [3, 4], even though it has been shown that, since an arbitrary number of very rarely observed objects might eventually be discovered, no point estimator can be generally unbiased in such infinite extrapolation [5, 6]. The genetics context requires a more modest extrapolation to a finite number of observations. In this case, the potential problem of an infinite number of rarely observed objects is eliminated: based on the random sampling assumption, we know that the rate of new discoveries per sample cannot increase with the number of samples. The number of discoveries can therefore be bounded with good confidence above and below. This does not guarantee the existence of an unbiased point estimator, but supports the idea that a jackknife estimator for finite extrapolation has the potential to be more reliable than one for infinite extrapolation. With this in mind, we write the expansion ˆV n N = V n + p a p i i N, n, i=1 S3 where N, n = N 1 j=n 1/j for a fixed jackknife order p. This assumption provides the expected behavior as n N, and is exact for p 1 in the case of a constant-size, neutrally evolving population. The parameters a p k are calculated by equating estimators ˆV n N = ˆV n 1 N = = ˆV n p N,
3 3 and solving the resulting p linear equations. Explicit expressions are given below. Figure S6 shows that, based on subsampled simulated data from our demographic model, our jackknife estimator could predict with good accuracy the number of segregating sites in the full data.. Parameters of the jackknife expansion The jackknife expansion parameters for the number of undiscovered variants in a sample of size N, based on the site frequency spectrum φi from a sample of size n, are easily derived using symbolic software using V n N = V n + p i=1 ap i i N, n, together with the equality ˆV n N = ˆV n 1 N = = ˆV n p N, and k k j N Φj =V n V n k j j=1 = p j=1 a p j j N, n k j N, n. S4 The resulting equations for the number of missed variants m p are bulky, but are simple rational expressions of n, N, and the φi. The predicted number m p of missing variants at second and third order are 1 + 5n 6n 2 + 2n 3 N, n n2 3n + n 2 N, n 2 m 2 = n3 5n + 2n 2 φ n 2 N, n n2 3n + n 2 N, n 2 + n3 5n + 2n 2 φ2 S5 at second order, and n + 885n 2 837n n 4 114n n 6 N, n m 3 = n n 637n n 3 18n n 5 φ n N, n2 5 51n + 81n 2 39n 3 + 6n n 2 3 4n + n 2 N, n n n + 281n 2 96n n 4 φ n + 61n n 3 195n n 5 6n 6 N, n 1 + n 2 n n + 281n 2 96n n 4 φ n 731n n n 4 51n 5 + 6n 6 N, n 2 n n 637n n 3 18n n 5 φ n n 22n 2 + 3n 3 N, n 3 n n + 281n 2 96n n 4 φ n3 N, n 3 + 2n + 5 8n + 3n 2 N, n 1 + n 2 n n 39n 2 + 6n 3 φ n3 2 + n N, n 3 n n 39n 2 + 6n 3 φ3 S6 at third order. limit of interest occurs when n >> 1, when the estimates simplify to m 1 = N, nφ1 m 2 = N, n + 2 N, n 2 φ1 2 N, nφ2 m 3 = N, n + 2 N, n + 3 N, n 2 6 φ1 3 N, n + 2 N, n φ2 + 3 N, nφ3. S7 Note that the number of terms of the site frequency spectrum that are used in the estimate is equal to the order of the jackknife expansion, and the magnitude of the coefficient of each SF S term increases with the perturbation order, in a way that may diverge for sufficiently large. s a result, for large values of, the jackknife expansion becomes unstable and sensitive to noise: the choice of the jackknife expansion order is therefore always a compromise.
4 4 B. Jackknife estimates for all exon pilot populations In figure S7, we show the jackknife projected number of sites to be discovered in all 7 populations from the exon pilot project. In Figure S6, we compare the results of the jacknife estimator applied to the expected SFS resulting from sampling 8 individuals from each of the three panels in our analysis. VI. LIKELIHOOD PROFILES ND BOOTSTRP DISTRIBUTION Figure S8 provides a comparison of bootstrap estimates to likelihood profiling. We provide the results in genetic units, as likelihood profiling requires holding parameters constant while optimizing other parameters, a task much simplified if carried in the units used in the simulation. Given N a and generation time g see Methods, the transformation from genetic to physical units is T Eus = 2N a gτ Eus T B = 2N a g τ Eus + τ B T f = 2N a g τ Eus + τ B + τ f N f = N a ν f N B = N a ν B N EU = N a ν EU N S = N a ν S r S = ν S /ν S g/t Eus 1 r EU = ν EU /ν EU g/t Eus 1 m i = M i /2N a. S8 s can be seen in Figure S8, likelihood profiles are in qualitative agreement with the bootstrap estimates. Bootstrap 95% confidence interval correspond roughly to the range 1-2 log-likelihood units in base e. Note that the profiles display composite likelihoods since linkage between neighboring sites in the Poisson Random Field approach creates correlations between neighboring sites. s a result, we expect that likelihood-based confidence intervals provide an underestimate of the true variance, better captured by the bootstrap analysis. [1] Harrow, J et al. 26 Gencode: producing a reference annotation for encode. Genome Biol 7, S [2] Hernandez, R. D, Williamson, S. H, & Bustamante, C. D. 27 Context dependence, ancestral misidentification, and spurious signatures of natural selection. Biol Evol 24, Mol [3] Burnham, K & Overton, W Estimation of the size of a closed population when capture probabilities vary among animals. Biometrika 65, [4] Burnham, K & Overton, W Robust estimation of population size when capture probabilities vary among animals. Ecology 6, [5] Link, W 23 Nonidentifiability of population size from capture-recapture data with heterogeneous detection probabilities Biometrics 59, [6] Link, W 26 Rejoinder to: "On identifiability in capture-recapture models". Biometrics 62,
5 FIG. S4. Two-dimensional marginal frequency spectra for a the low-coverage pilot, b the exon pilot, and d the exon pilot once the error model has been applied. c shows the nscombe residuals between the two pilots, whereas e shows the residuals after the error model has been applied to the exon pilot. ll spectra are polarized using the March 26 assembly of the chimp genome PanTro2. 5
6 6 number of sites CEU, model CEU, data CHB+JPT, model CHB+JPT, data YRI, model YRI, data derived allele counts out of 8 FIG. S5. Comparison of model predictions, based on our maximumlikelihood parameters, and data from 4-fold synonymous sites polarized using chimp as an outgroup. The discrepancy for very common variants is due to cases where the chimp allele is not the ancestral allele [2]. Segregating sites x True Obs. Proj. YRI CEU CHBJPT Sample size 2x diploid sample size FIG. S6. To control for bias of the jackknife estimator, we used our demographic model to generate "True" SFS for 1 chromosomes in our 3 population panels. We then calculated average jackknife projections based on "observed" subsamples of 8 chromosomes for each population. In the absence of sampling, we find little bias for the first decade of extrapolation. Counts in Exon pilot region Obs. Proj. CEU TSI CHB CHD JPT LWK YRI Sample size 2x diploid sample size FIG. S7. Observations and third-order jackknife predictions of the number of variants to be discovered in the exon capture panels as sample size is increased, based on 8 chromosomes per population.
7 FIG. S8. Bootstrap and likelihood profiles for the Out-Of-frica model for parameters expressed in genetic units. Likelihood profiles are obtained in the usual way by fixing one parameter and optimizing the likelihoods over the other parameters. 7
Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Information #
 Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Details of PRF Methodology In the Poisson Random Field PRF) model, it is assumed that non-synonymous mutations at a given gene are either
Bustamante et al., Supplementary Nature Manuscript # 1 out of 9 Details of PRF Methodology In the Poisson Random Field PRF) model, it is assumed that non-synonymous mutations at a given gene are either
Frequency Spectra and Inference in Population Genetics
 Frequency Spectra and Inference in Population Genetics Although coalescent models have come to play a central role in population genetics, there are some situations where genealogies may not lead to efficient
Frequency Spectra and Inference in Population Genetics Although coalescent models have come to play a central role in population genetics, there are some situations where genealogies may not lead to efficient
Robust demographic inference from genomic and SNP data
 Robust demographic inference from genomic and SNP data Laurent Excoffier Isabelle Duperret, Emilia Huerta-Sanchez, Matthieu Foll, Vitor Sousa, Isabel Alves Computational and Molecular Population Genetics
Robust demographic inference from genomic and SNP data Laurent Excoffier Isabelle Duperret, Emilia Huerta-Sanchez, Matthieu Foll, Vitor Sousa, Isabel Alves Computational and Molecular Population Genetics
Supporting Information Text S1
 Supporting Information Text S1 List of Supplementary Figures S1 The fraction of SNPs s where there is an excess of Neandertal derived alleles n over Denisova derived alleles d as a function of the derived
Supporting Information Text S1 List of Supplementary Figures S1 The fraction of SNPs s where there is an excess of Neandertal derived alleles n over Denisova derived alleles d as a function of the derived
SWEEPFINDER2: Increased sensitivity, robustness, and flexibility
 SWEEPFINDER2: Increased sensitivity, robustness, and flexibility Michael DeGiorgio 1,*, Christian D. Huber 2, Melissa J. Hubisz 3, Ines Hellmann 4, and Rasmus Nielsen 5 1 Department of Biology, Pennsylvania
SWEEPFINDER2: Increased sensitivity, robustness, and flexibility Michael DeGiorgio 1,*, Christian D. Huber 2, Melissa J. Hubisz 3, Ines Hellmann 4, and Rasmus Nielsen 5 1 Department of Biology, Pennsylvania
Supporting Information
 Supporting Information Hammer et al. 10.1073/pnas.1109300108 SI Materials and Methods Two-Population Model. Estimating demographic parameters. For each pair of sub-saharan African populations we consider
Supporting Information Hammer et al. 10.1073/pnas.1109300108 SI Materials and Methods Two-Population Model. Estimating demographic parameters. For each pair of sub-saharan African populations we consider
The Wright-Fisher Model and Genetic Drift
 The Wright-Fisher Model and Genetic Drift January 22, 2015 1 1 Hardy-Weinberg Equilibrium Our goal is to understand the dynamics of allele and genotype frequencies in an infinite, randomlymating population
The Wright-Fisher Model and Genetic Drift January 22, 2015 1 1 Hardy-Weinberg Equilibrium Our goal is to understand the dynamics of allele and genotype frequencies in an infinite, randomlymating population
Figure S1: The model underlying our inference of the age of ancient genomes
 A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years Priya Moorjani, Sriram Sankararaman, Qiaomei Fu, Molly Przeworski, Nick Patterson,
A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years Priya Moorjani, Sriram Sankararaman, Qiaomei Fu, Molly Przeworski, Nick Patterson,
Introduction to Natural Selection. Ryan Hernandez Tim O Connor
 Introduction to Natural Selection Ryan Hernandez Tim O Connor 1 Goals Learn about the population genetics of natural selection How to write a simple simulation with natural selection 2 Basic Biology genome
Introduction to Natural Selection Ryan Hernandez Tim O Connor 1 Goals Learn about the population genetics of natural selection How to write a simple simulation with natural selection 2 Basic Biology genome
Genotype Imputation. Class Discussion for January 19, 2016
 Genotype Imputation Class Discussion for January 19, 2016 Intuition Patterns of genetic variation in one individual guide our interpretation of the genomes of other individuals Imputation uses previously
Genotype Imputation Class Discussion for January 19, 2016 Intuition Patterns of genetic variation in one individual guide our interpretation of the genomes of other individuals Imputation uses previously
Package SPECIES. R topics documented: April 23, Type Package. Title Statistical package for species richness estimation. Version 1.
 Package SPECIES April 23, 2011 Type Package Title Statistical package for species richness estimation Version 1.0 Date 2010-01-24 Author Ji-Ping Wang, Maintainer Ji-Ping Wang
Package SPECIES April 23, 2011 Type Package Title Statistical package for species richness estimation Version 1.0 Date 2010-01-24 Author Ji-Ping Wang, Maintainer Ji-Ping Wang
Estimating Evolutionary Trees. Phylogenetic Methods
 Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
A Practitioner s Guide to Cluster-Robust Inference
 A Practitioner s Guide to Cluster-Robust Inference A. C. Cameron and D. L. Miller presented by Federico Curci March 4, 2015 Cameron Miller Cluster Clinic II March 4, 2015 1 / 20 In the previous episode
A Practitioner s Guide to Cluster-Robust Inference A. C. Cameron and D. L. Miller presented by Federico Curci March 4, 2015 Cameron Miller Cluster Clinic II March 4, 2015 1 / 20 In the previous episode
CSci 8980: Advanced Topics in Graphical Models Analysis of Genetic Variation
 CSci 8980: Advanced Topics in Graphical Models Analysis of Genetic Variation Instructor: Arindam Banerjee November 26, 2007 Genetic Polymorphism Single nucleotide polymorphism (SNP) Genetic Polymorphism
CSci 8980: Advanced Topics in Graphical Models Analysis of Genetic Variation Instructor: Arindam Banerjee November 26, 2007 Genetic Polymorphism Single nucleotide polymorphism (SNP) Genetic Polymorphism
Haplotype-based variant detection from short-read sequencing
 Haplotype-based variant detection from short-read sequencing Erik Garrison and Gabor Marth July 16, 2012 1 Motivation While statistical phasing approaches are necessary for the determination of large-scale
Haplotype-based variant detection from short-read sequencing Erik Garrison and Gabor Marth July 16, 2012 1 Motivation While statistical phasing approaches are necessary for the determination of large-scale
THE N-VALUE GAME OVER Z AND R
 THE N-VALUE GAME OVER Z AND R YIDA GAO, MATT REDMOND, ZACH STEWARD Abstract. The n-value game is an easily described mathematical diversion with deep underpinnings in dynamical systems analysis. We examine
THE N-VALUE GAME OVER Z AND R YIDA GAO, MATT REDMOND, ZACH STEWARD Abstract. The n-value game is an easily described mathematical diversion with deep underpinnings in dynamical systems analysis. We examine
Genetic Drift in Human Evolution
 Genetic Drift in Human Evolution (Part 2 of 2) 1 Ecology and Evolutionary Biology Center for Computational Molecular Biology Brown University Outline Introduction to genetic drift Modeling genetic drift
Genetic Drift in Human Evolution (Part 2 of 2) 1 Ecology and Evolutionary Biology Center for Computational Molecular Biology Brown University Outline Introduction to genetic drift Modeling genetic drift
Association studies and regression
 Association studies and regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Association studies and regression 1 / 104 Administration
Association studies and regression CM226: Machine Learning for Bioinformatics. Fall 2016 Sriram Sankararaman Acknowledgments: Fei Sha, Ameet Talwalkar Association studies and regression 1 / 104 Administration
Diffusion Models in Population Genetics
 Diffusion Models in Population Genetics Laura Kubatko kubatko.2@osu.edu MBI Workshop on Spatially-varying stochastic differential equations, with application to the biological sciences July 10, 2015 Laura
Diffusion Models in Population Genetics Laura Kubatko kubatko.2@osu.edu MBI Workshop on Spatially-varying stochastic differential equations, with application to the biological sciences July 10, 2015 Laura
Multilevel Statistical Models: 3 rd edition, 2003 Contents
 Multilevel Statistical Models: 3 rd edition, 2003 Contents Preface Acknowledgements Notation Two and three level models. A general classification notation and diagram Glossary Chapter 1 An introduction
Multilevel Statistical Models: 3 rd edition, 2003 Contents Preface Acknowledgements Notation Two and three level models. A general classification notation and diagram Glossary Chapter 1 An introduction
Pyrobayes: an improved base caller for SNP discovery in pyrosequences
 Pyrobayes: an improved base caller for SNP discovery in pyrosequences Aaron R Quinlan, Donald A Stewart, Michael P Strömberg & Gábor T Marth Supplementary figures and text: Supplementary Figure 1. The
Pyrobayes: an improved base caller for SNP discovery in pyrosequences Aaron R Quinlan, Donald A Stewart, Michael P Strömberg & Gábor T Marth Supplementary figures and text: Supplementary Figure 1. The
When is selection effective?
 Genetics: Early Online, published on March 23, 2016 as 10.153/genetics.115.18630 GENETICS INVESTIGATION When is selection effective? Simon Gravel,1 McGill University, Montreal, Canada ABSTRACT Deleterious
Genetics: Early Online, published on March 23, 2016 as 10.153/genetics.115.18630 GENETICS INVESTIGATION When is selection effective? Simon Gravel,1 McGill University, Montreal, Canada ABSTRACT Deleterious
Effect of Genetic Divergence in Identifying Ancestral Origin using HAPAA
 Effect of Genetic Divergence in Identifying Ancestral Origin using HAPAA Andreas Sundquist*, Eugene Fratkin*, Chuong B. Do, Serafim Batzoglou Department of Computer Science, Stanford University, Stanford,
Effect of Genetic Divergence in Identifying Ancestral Origin using HAPAA Andreas Sundquist*, Eugene Fratkin*, Chuong B. Do, Serafim Batzoglou Department of Computer Science, Stanford University, Stanford,
Lecture 22: Signatures of Selection and Introduction to Linkage Disequilibrium. November 12, 2012
 Lecture 22: Signatures of Selection and Introduction to Linkage Disequilibrium November 12, 2012 Last Time Sequence data and quantification of variation Infinite sites model Nucleotide diversity (π) Sequence-based
Lecture 22: Signatures of Selection and Introduction to Linkage Disequilibrium November 12, 2012 Last Time Sequence data and quantification of variation Infinite sites model Nucleotide diversity (π) Sequence-based
1. Understand the methods for analyzing population structure in genomes
 MSCBIO 2070/02-710: Computational Genomics, Spring 2016 HW3: Population Genetics Due: 24:00 EST, April 4, 2016 by autolab Your goals in this assignment are to 1. Understand the methods for analyzing population
MSCBIO 2070/02-710: Computational Genomics, Spring 2016 HW3: Population Genetics Due: 24:00 EST, April 4, 2016 by autolab Your goals in this assignment are to 1. Understand the methods for analyzing population
Food delivered. Food obtained S 3
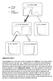 Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Population genetics models of local ancestry
 Genetics: Published Articles Ahead of Print, published on April 4, 2012 as 10.1534/genetics.112.139808 Population genetics models of local ancestry Simon Gravel Genetics Department Stanford University
Genetics: Published Articles Ahead of Print, published on April 4, 2012 as 10.1534/genetics.112.139808 Population genetics models of local ancestry Simon Gravel Genetics Department Stanford University
Mark-recapture with identification errors
 Mark-recapture with identification errors Richard Vale 16/07/14 1 Hard to estimate the size of an animal population. One popular method: markrecapture sampling 2 A population A sample, taken by a biologist
Mark-recapture with identification errors Richard Vale 16/07/14 1 Hard to estimate the size of an animal population. One popular method: markrecapture sampling 2 A population A sample, taken by a biologist
A better way to bootstrap pairs
 A better way to bootstrap pairs Emmanuel Flachaire GREQAM - Université de la Méditerranée CORE - Université Catholique de Louvain April 999 Abstract In this paper we are interested in heteroskedastic regression
A better way to bootstrap pairs Emmanuel Flachaire GREQAM - Université de la Méditerranée CORE - Université Catholique de Louvain April 999 Abstract In this paper we are interested in heteroskedastic regression
Algorithm-Independent Learning Issues
 Algorithm-Independent Learning Issues Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2007 c 2007, Selim Aksoy Introduction We have seen many learning
Algorithm-Independent Learning Issues Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2007 c 2007, Selim Aksoy Introduction We have seen many learning
Genotype Imputation. Biostatistics 666
 Genotype Imputation Biostatistics 666 Previously Hidden Markov Models for Relative Pairs Linkage analysis using affected sibling pairs Estimation of pairwise relationships Identity-by-Descent Relatives
Genotype Imputation Biostatistics 666 Previously Hidden Markov Models for Relative Pairs Linkage analysis using affected sibling pairs Estimation of pairwise relationships Identity-by-Descent Relatives
Phylogenetics: Bayesian Phylogenetic Analysis. COMP Spring 2015 Luay Nakhleh, Rice University
 Phylogenetics: Bayesian Phylogenetic Analysis COMP 571 - Spring 2015 Luay Nakhleh, Rice University Bayes Rule P(X = x Y = y) = P(X = x, Y = y) P(Y = y) = P(X = x)p(y = y X = x) P x P(X = x 0 )P(Y = y X
Phylogenetics: Bayesian Phylogenetic Analysis COMP 571 - Spring 2015 Luay Nakhleh, Rice University Bayes Rule P(X = x Y = y) = P(X = x, Y = y) P(Y = y) = P(X = x)p(y = y X = x) P x P(X = x 0 )P(Y = y X
Linear Regression (1/1/17)
 STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
STA613/CBB540: Statistical methods in computational biology Linear Regression (1/1/17) Lecturer: Barbara Engelhardt Scribe: Ethan Hada 1. Linear regression 1.1. Linear regression basics. Linear regression
Fitness landscapes and seascapes
 Fitness landscapes and seascapes Michael Lässig Institute for Theoretical Physics University of Cologne Thanks Ville Mustonen: Cross-species analysis of bacterial promoters, Nonequilibrium evolution of
Fitness landscapes and seascapes Michael Lässig Institute for Theoretical Physics University of Cologne Thanks Ville Mustonen: Cross-species analysis of bacterial promoters, Nonequilibrium evolution of
A Simulation Comparison Study for Estimating the Process Capability Index C pm with Asymmetric Tolerances
 Available online at ijims.ms.tku.edu.tw/list.asp International Journal of Information and Management Sciences 20 (2009), 243-253 A Simulation Comparison Study for Estimating the Process Capability Index
Available online at ijims.ms.tku.edu.tw/list.asp International Journal of Information and Management Sciences 20 (2009), 243-253 A Simulation Comparison Study for Estimating the Process Capability Index
A8824: Statistics Notes David Weinberg, Astronomy 8824: Statistics Notes 6 Estimating Errors From Data
 Where does the error bar go? Astronomy 8824: Statistics otes 6 Estimating Errors From Data Suppose you measure the average depression of flux in a quasar caused by absorption from the Lyman-alpha forest.
Where does the error bar go? Astronomy 8824: Statistics otes 6 Estimating Errors From Data Suppose you measure the average depression of flux in a quasar caused by absorption from the Lyman-alpha forest.
Detecting selection from differentiation between populations: the FLK and hapflk approach.
 Detecting selection from differentiation between populations: the FLK and hapflk approach. Bertrand Servin bservin@toulouse.inra.fr Maria-Ines Fariello, Simon Boitard, Claude Chevalet, Magali SanCristobal,
Detecting selection from differentiation between populations: the FLK and hapflk approach. Bertrand Servin bservin@toulouse.inra.fr Maria-Ines Fariello, Simon Boitard, Claude Chevalet, Magali SanCristobal,
Figure S2. The distribution of the sizes (in bp) of syntenic regions of humans and chimpanzees on human chromosome 21.
 Frequency 0 1000 2000 3000 4000 5000 0 2 4 6 8 10 Distance Figure S1. The distribution of human-chimpanzee sequence divergence for syntenic regions of humans and chimpanzees on human chromosome 21. Distance
Frequency 0 1000 2000 3000 4000 5000 0 2 4 6 8 10 Distance Figure S1. The distribution of human-chimpanzee sequence divergence for syntenic regions of humans and chimpanzees on human chromosome 21. Distance
Phylogenetic inference
 Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Intensive Course on Population Genetics. Ville Mustonen
 Intensive Course on Population Genetics Ville Mustonen Population genetics: The study of the distribution of inherited variation among a group of organisms of the same species [Oxford Dictionary of Biology]
Intensive Course on Population Genetics Ville Mustonen Population genetics: The study of the distribution of inherited variation among a group of organisms of the same species [Oxford Dictionary of Biology]
Bayesian Inference using Markov Chain Monte Carlo in Phylogenetic Studies
 Bayesian Inference using Markov Chain Monte Carlo in Phylogenetic Studies 1 What is phylogeny? Essay written for the course in Markov Chains 2004 Torbjörn Karfunkel Phylogeny is the evolutionary development
Bayesian Inference using Markov Chain Monte Carlo in Phylogenetic Studies 1 What is phylogeny? Essay written for the course in Markov Chains 2004 Torbjörn Karfunkel Phylogeny is the evolutionary development
Inferring Admixture Histories of Human Populations Using. Linkage Disequilibrium
 Genetics: Early Online, published on February 14, 2013 as 10.1534/genetics.112.147330 Inferring Admixture Histories of Human Populations Using Linkage Disequilibrium Po-Ru Loh,1, Mark Lipson,1, Nick Patterson,
Genetics: Early Online, published on February 14, 2013 as 10.1534/genetics.112.147330 Inferring Admixture Histories of Human Populations Using Linkage Disequilibrium Po-Ru Loh,1, Mark Lipson,1, Nick Patterson,
Introduction to Survey Data Analysis
 Introduction to Survey Data Analysis JULY 2011 Afsaneh Yazdani Preface Learning from Data Four-step process by which we can learn from data: 1. Defining the Problem 2. Collecting the Data 3. Summarizing
Introduction to Survey Data Analysis JULY 2011 Afsaneh Yazdani Preface Learning from Data Four-step process by which we can learn from data: 1. Defining the Problem 2. Collecting the Data 3. Summarizing
Basic Sampling Methods
 Basic Sampling Methods Sargur Srihari srihari@cedar.buffalo.edu 1 1. Motivation Topics Intractability in ML How sampling can help 2. Ancestral Sampling Using BNs 3. Transforming a Uniform Distribution
Basic Sampling Methods Sargur Srihari srihari@cedar.buffalo.edu 1 1. Motivation Topics Intractability in ML How sampling can help 2. Ancestral Sampling Using BNs 3. Transforming a Uniform Distribution
Degrees-of-freedom estimation in the Student-t noise model.
 LIGO-T0497 Degrees-of-freedom estimation in the Student-t noise model. Christian Röver September, 0 Introduction The Student-t noise model was introduced in [] as a robust alternative to the commonly used
LIGO-T0497 Degrees-of-freedom estimation in the Student-t noise model. Christian Röver September, 0 Introduction The Student-t noise model was introduced in [] as a robust alternative to the commonly used
RandNLA: Randomization in Numerical Linear Algebra
 RandNLA: Randomization in Numerical Linear Algebra Petros Drineas Department of Computer Science Rensselaer Polytechnic Institute To access my web page: drineas Why RandNLA? Randomization and sampling
RandNLA: Randomization in Numerical Linear Algebra Petros Drineas Department of Computer Science Rensselaer Polytechnic Institute To access my web page: drineas Why RandNLA? Randomization and sampling
Empirical Bayes Moderation of Asymptotically Linear Parameters
 Empirical Bayes Moderation of Asymptotically Linear Parameters Nima Hejazi Division of Biostatistics University of California, Berkeley stat.berkeley.edu/~nhejazi nimahejazi.org twitter/@nshejazi github/nhejazi
Empirical Bayes Moderation of Asymptotically Linear Parameters Nima Hejazi Division of Biostatistics University of California, Berkeley stat.berkeley.edu/~nhejazi nimahejazi.org twitter/@nshejazi github/nhejazi
Estimation of the Conditional Variance in Paired Experiments
 Estimation of the Conditional Variance in Paired Experiments Alberto Abadie & Guido W. Imbens Harvard University and BER June 008 Abstract In paired randomized experiments units are grouped in pairs, often
Estimation of the Conditional Variance in Paired Experiments Alberto Abadie & Guido W. Imbens Harvard University and BER June 008 Abstract In paired randomized experiments units are grouped in pairs, often
SEQUENCE DIVERGENCE,FUNCTIONAL CONSTRAINT, AND SELECTION IN PROTEIN EVOLUTION
 Annu. Rev. Genomics Hum. Genet. 2003. 4:213 35 doi: 10.1146/annurev.genom.4.020303.162528 Copyright c 2003 by Annual Reviews. All rights reserved First published online as a Review in Advance on June 4,
Annu. Rev. Genomics Hum. Genet. 2003. 4:213 35 doi: 10.1146/annurev.genom.4.020303.162528 Copyright c 2003 by Annual Reviews. All rights reserved First published online as a Review in Advance on June 4,
Chapter 1 Statistical Inference
 Chapter 1 Statistical Inference causal inference To infer causality, you need a randomized experiment (or a huge observational study and lots of outside information). inference to populations Generalizations
Chapter 1 Statistical Inference causal inference To infer causality, you need a randomized experiment (or a huge observational study and lots of outside information). inference to populations Generalizations
122 9 NEUTRALITY TESTS
 122 9 NEUTRALITY TESTS 9 Neutrality Tests Up to now, we calculated different things from various models and compared our findings with data. But to be able to state, with some quantifiable certainty, that
122 9 NEUTRALITY TESTS 9 Neutrality Tests Up to now, we calculated different things from various models and compared our findings with data. But to be able to state, with some quantifiable certainty, that
7. Tests for selection
 Sequence analysis and genomics 7. Tests for selection Dr. Katja Nowick Group leader TFome and Transcriptome Evolution Bioinformatics group Paul-Flechsig-Institute for Brain Research www. nowicklab.info
Sequence analysis and genomics 7. Tests for selection Dr. Katja Nowick Group leader TFome and Transcriptome Evolution Bioinformatics group Paul-Flechsig-Institute for Brain Research www. nowicklab.info
Consistency Index (CI)
 Consistency Index (CI) minimum number of changes divided by the number required on the tree. CI=1 if there is no homoplasy negatively correlated with the number of species sampled Retention Index (RI)
Consistency Index (CI) minimum number of changes divided by the number required on the tree. CI=1 if there is no homoplasy negatively correlated with the number of species sampled Retention Index (RI)
Drosophila melanogaster and D. simulans, two fruit fly species that are nearly
 Comparative Genomics: Human versus chimpanzee 1. Introduction The chimpanzee is the closest living relative to humans. The two species are nearly identical in DNA sequence (>98% identity), yet vastly different
Comparative Genomics: Human versus chimpanzee 1. Introduction The chimpanzee is the closest living relative to humans. The two species are nearly identical in DNA sequence (>98% identity), yet vastly different
Statistical Data Analysis Stat 3: p-values, parameter estimation
 Statistical Data Analysis Stat 3: p-values, parameter estimation London Postgraduate Lectures on Particle Physics; University of London MSci course PH4515 Glen Cowan Physics Department Royal Holloway,
Statistical Data Analysis Stat 3: p-values, parameter estimation London Postgraduate Lectures on Particle Physics; University of London MSci course PH4515 Glen Cowan Physics Department Royal Holloway,
Bayesian Interpretations of Heteroskedastic Consistent Covariance Estimators Using the Informed Bayesian Bootstrap
 Bayesian Interpretations of Heteroskedastic Consistent Covariance Estimators Using the Informed Bayesian Bootstrap Dale J. Poirier University of California, Irvine September 1, 2008 Abstract This paper
Bayesian Interpretations of Heteroskedastic Consistent Covariance Estimators Using the Informed Bayesian Bootstrap Dale J. Poirier University of California, Irvine September 1, 2008 Abstract This paper
Genetic Variation in Finite Populations
 Genetic Variation in Finite Populations The amount of genetic variation found in a population is influenced by two opposing forces: mutation and genetic drift. 1 Mutation tends to increase variation. 2
Genetic Variation in Finite Populations The amount of genetic variation found in a population is influenced by two opposing forces: mutation and genetic drift. 1 Mutation tends to increase variation. 2
Phylogenetics. Applications of phylogenetics. Unrooted networks vs. rooted trees. Outline
 Phylogenetics Todd Vision iology 522 March 26, 2007 pplications of phylogenetics Studying organismal or biogeographic history Systematics ating events in the fossil record onservation biology Studying
Phylogenetics Todd Vision iology 522 March 26, 2007 pplications of phylogenetics Studying organismal or biogeographic history Systematics ating events in the fossil record onservation biology Studying
Recombination rate estimation in the presence of hotspots
 Methods Recombination rate estimation in the presence of hotspots Adam Auton 1 and Gil McVean Department of Statistics, University of Oxford, Oxford OX1 3TG, United Kingdom Fine-scale estimation of recombination
Methods Recombination rate estimation in the presence of hotspots Adam Auton 1 and Gil McVean Department of Statistics, University of Oxford, Oxford OX1 3TG, United Kingdom Fine-scale estimation of recombination
Marginal Screening and Post-Selection Inference
 Marginal Screening and Post-Selection Inference Ian McKeague August 13, 2017 Ian McKeague (Columbia University) Marginal Screening August 13, 2017 1 / 29 Outline 1 Background on Marginal Screening 2 2
Marginal Screening and Post-Selection Inference Ian McKeague August 13, 2017 Ian McKeague (Columbia University) Marginal Screening August 13, 2017 1 / 29 Outline 1 Background on Marginal Screening 2 2
Journal of Statistical Software
 JSS Journal of Statistical Software April 2011, Volume 40, Issue 9. http://www.jstatsoft.org/ SPECIES: An R Package for Species Richness Estimation Ji-Ping Wang Northwestern University Abstract We introduce
JSS Journal of Statistical Software April 2011, Volume 40, Issue 9. http://www.jstatsoft.org/ SPECIES: An R Package for Species Richness Estimation Ji-Ping Wang Northwestern University Abstract We introduce
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics
 POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
Previous lecture. Single variant association. Use genome-wide SNPs to account for confounding (population substructure)
 Previous lecture Single variant association Use genome-wide SNPs to account for confounding (population substructure) Estimation of effect size and winner s curse Meta-Analysis Today s outline P-value
Previous lecture Single variant association Use genome-wide SNPs to account for confounding (population substructure) Estimation of effect size and winner s curse Meta-Analysis Today s outline P-value
Fast and robust bootstrap for LTS
 Fast and robust bootstrap for LTS Gert Willems a,, Stefan Van Aelst b a Department of Mathematics and Computer Science, University of Antwerp, Middelheimlaan 1, B-2020 Antwerp, Belgium b Department of
Fast and robust bootstrap for LTS Gert Willems a,, Stefan Van Aelst b a Department of Mathematics and Computer Science, University of Antwerp, Middelheimlaan 1, B-2020 Antwerp, Belgium b Department of
Why is the field of statistics still an active one?
 Why is the field of statistics still an active one? It s obvious that one needs statistics: to describe experimental data in a compact way, to compare datasets, to ask whether data are consistent with
Why is the field of statistics still an active one? It s obvious that one needs statistics: to describe experimental data in a compact way, to compare datasets, to ask whether data are consistent with
6 Introduction to Population Genetics
 70 Grundlagen der Bioinformatik, SoSe 11, D. Huson, May 19, 2011 6 Introduction to Population Genetics This chapter is based on: J. Hein, M.H. Schierup and C. Wuif, Gene genealogies, variation and evolution,
70 Grundlagen der Bioinformatik, SoSe 11, D. Huson, May 19, 2011 6 Introduction to Population Genetics This chapter is based on: J. Hein, M.H. Schierup and C. Wuif, Gene genealogies, variation and evolution,
On robust and efficient estimation of the center of. Symmetry.
 On robust and efficient estimation of the center of symmetry Howard D. Bondell Department of Statistics, North Carolina State University Raleigh, NC 27695-8203, U.S.A (email: bondell@stat.ncsu.edu) Abstract
On robust and efficient estimation of the center of symmetry Howard D. Bondell Department of Statistics, North Carolina State University Raleigh, NC 27695-8203, U.S.A (email: bondell@stat.ncsu.edu) Abstract
CHAO, JACKKNIFE AND BOOTSTRAP ESTIMATORS OF SPECIES RICHNESS
 IJAMAA, Vol. 12, No. 1, (January-June 2017), pp. 7-15 Serials Publications ISSN: 0973-3868 CHAO, JACKKNIFE AND BOOTSTRAP ESTIMATORS OF SPECIES RICHNESS CHAVAN KR. SARMAH ABSTRACT: The species richness
IJAMAA, Vol. 12, No. 1, (January-June 2017), pp. 7-15 Serials Publications ISSN: 0973-3868 CHAO, JACKKNIFE AND BOOTSTRAP ESTIMATORS OF SPECIES RICHNESS CHAVAN KR. SARMAH ABSTRACT: The species richness
Selection and Population Genetics
 Selection and Population Genetics Evolution by natural selection can occur when three conditions are satisfied: Variation within populations - individuals have different traits (phenotypes). height and
Selection and Population Genetics Evolution by natural selection can occur when three conditions are satisfied: Variation within populations - individuals have different traits (phenotypes). height and
if n is large, Z i are weakly dependent 0-1-variables, p i = P(Z i = 1) small, and Then n approx i=1 i=1 n i=1
 Count models A classical, theoretical argument for the Poisson distribution is the approximation Binom(n, p) Pois(λ) for large n and small p and λ = np. This can be extended considerably to n approx Z
Count models A classical, theoretical argument for the Poisson distribution is the approximation Binom(n, p) Pois(λ) for large n and small p and λ = np. This can be extended considerably to n approx Z
Introduction to Advanced Population Genetics
 Introduction to Advanced Population Genetics Learning Objectives Describe the basic model of human evolutionary history Describe the key evolutionary forces How demography can influence the site frequency
Introduction to Advanced Population Genetics Learning Objectives Describe the basic model of human evolutionary history Describe the key evolutionary forces How demography can influence the site frequency
A (short) introduction to phylogenetics
 A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
What s New in Econometrics. Lecture 1
 What s New in Econometrics Lecture 1 Estimation of Average Treatment Effects Under Unconfoundedness Guido Imbens NBER Summer Institute, 2007 Outline 1. Introduction 2. Potential Outcomes 3. Estimands and
What s New in Econometrics Lecture 1 Estimation of Average Treatment Effects Under Unconfoundedness Guido Imbens NBER Summer Institute, 2007 Outline 1. Introduction 2. Potential Outcomes 3. Estimands and
Modelling Linkage Disequilibrium, And Identifying Recombination Hotspots Using SNP Data
 Modelling Linkage Disequilibrium, And Identifying Recombination Hotspots Using SNP Data Na Li and Matthew Stephens July 25, 2003 Department of Biostatistics, University of Washington, Seattle, WA 98195
Modelling Linkage Disequilibrium, And Identifying Recombination Hotspots Using SNP Data Na Li and Matthew Stephens July 25, 2003 Department of Biostatistics, University of Washington, Seattle, WA 98195
Learning ancestral genetic processes using nonparametric Bayesian models
 Learning ancestral genetic processes using nonparametric Bayesian models Kyung-Ah Sohn October 31, 2011 Committee Members: Eric P. Xing, Chair Zoubin Ghahramani Russell Schwartz Kathryn Roeder Matthew
Learning ancestral genetic processes using nonparametric Bayesian models Kyung-Ah Sohn October 31, 2011 Committee Members: Eric P. Xing, Chair Zoubin Ghahramani Russell Schwartz Kathryn Roeder Matthew
A comparison of two popular statistical methods for estimating the time to most recent common ancestor (TMRCA) from a sample of DNA sequences
 Indian Academy of Sciences A comparison of two popular statistical methods for estimating the time to most recent common ancestor (TMRCA) from a sample of DNA sequences ANALABHA BASU and PARTHA P. MAJUMDER*
Indian Academy of Sciences A comparison of two popular statistical methods for estimating the time to most recent common ancestor (TMRCA) from a sample of DNA sequences ANALABHA BASU and PARTHA P. MAJUMDER*
The abundance of deleterious polymorphisms in humans
 Genetics: Published Articles Ahead of Print, published on February 23, 2012 as 10.1534/genetics.111.137893 Note February 3, 2011 The abundance of deleterious polymorphisms in humans Sankar Subramanian
Genetics: Published Articles Ahead of Print, published on February 23, 2012 as 10.1534/genetics.111.137893 Note February 3, 2011 The abundance of deleterious polymorphisms in humans Sankar Subramanian
Bootstrap, Jackknife and other resampling methods
 Bootstrap, Jackknife and other resampling methods Part VI: Cross-validation Rozenn Dahyot Room 128, Department of Statistics Trinity College Dublin, Ireland dahyot@mee.tcd.ie 2005 R. Dahyot (TCD) 453 Modern
Bootstrap, Jackknife and other resampling methods Part VI: Cross-validation Rozenn Dahyot Room 128, Department of Statistics Trinity College Dublin, Ireland dahyot@mee.tcd.ie 2005 R. Dahyot (TCD) 453 Modern
BFF Four: Are we Converging?
 BFF Four: Are we Converging? Nancy Reid May 2, 2017 Classical Approaches: A Look Way Back Nature of Probability BFF one to three: a look back Comparisons Are we getting there? BFF Four Harvard, May 2017
BFF Four: Are we Converging? Nancy Reid May 2, 2017 Classical Approaches: A Look Way Back Nature of Probability BFF one to three: a look back Comparisons Are we getting there? BFF Four Harvard, May 2017
Site Occupancy Models with Heterogeneous Detection Probabilities
 Biometrics 62, 97 102 March 2006 DOI: 10.1111/j.1541-0420.2005.00439.x Site Occupancy Models with Heterogeneous Detection Probabilities J. Andrew Royle USGS Patuxent Wildlife Research Center, 12100 Beech
Biometrics 62, 97 102 March 2006 DOI: 10.1111/j.1541-0420.2005.00439.x Site Occupancy Models with Heterogeneous Detection Probabilities J. Andrew Royle USGS Patuxent Wildlife Research Center, 12100 Beech
6 Introduction to Population Genetics
 Grundlagen der Bioinformatik, SoSe 14, D. Huson, May 18, 2014 67 6 Introduction to Population Genetics This chapter is based on: J. Hein, M.H. Schierup and C. Wuif, Gene genealogies, variation and evolution,
Grundlagen der Bioinformatik, SoSe 14, D. Huson, May 18, 2014 67 6 Introduction to Population Genetics This chapter is based on: J. Hein, M.H. Schierup and C. Wuif, Gene genealogies, variation and evolution,
Four aspects of a sampling strategy necessary to make accurate and precise inferences about populations are:
 Why Sample? Often researchers are interested in answering questions about a particular population. They might be interested in the density, species richness, or specific life history parameters such as
Why Sample? Often researchers are interested in answering questions about a particular population. They might be interested in the density, species richness, or specific life history parameters such as
De novo assembly and genotyping of variants using colored de Bruijn graphs
 De novo assembly and genotyping of variants using colored de Bruijn graphs Iqbal et al. 2012 Kolmogorov Mikhail 2013 Challenges Detecting genetic variants that are highly divergent from a reference Detecting
De novo assembly and genotyping of variants using colored de Bruijn graphs Iqbal et al. 2012 Kolmogorov Mikhail 2013 Challenges Detecting genetic variants that are highly divergent from a reference Detecting
Econometrics Spring School 2016 Econometric Modelling. Lecture 6: Model selection theory and evidence Introduction to Monte Carlo Simulation
 Econometrics Spring School 2016 Econometric Modelling Jurgen A Doornik, David F. Hendry, and Felix Pretis George-Washington University March 2016 Lecture 6: Model selection theory and evidence Introduction
Econometrics Spring School 2016 Econometric Modelling Jurgen A Doornik, David F. Hendry, and Felix Pretis George-Washington University March 2016 Lecture 6: Model selection theory and evidence Introduction
p(d g A,g B )p(g B ), g B
 Supplementary Note Marginal effects for two-locus models Here we derive the marginal effect size of the three models given in Figure 1 of the main text. For each model we assume the two loci (A and B)
Supplementary Note Marginal effects for two-locus models Here we derive the marginal effect size of the three models given in Figure 1 of the main text. For each model we assume the two loci (A and B)
Estimating population size by spatially explicit capture recapture
 Estimating population size by spatially explicit capture recapture Murray G. Efford 1 and Rachel M. Fewster 2 1. 60 Helensburgh Road, Dunedin 9010, New Zealand. murray.efford@gmail.com Ph +64 3 476 4668.
Estimating population size by spatially explicit capture recapture Murray G. Efford 1 and Rachel M. Fewster 2 1. 60 Helensburgh Road, Dunedin 9010, New Zealand. murray.efford@gmail.com Ph +64 3 476 4668.
When Should We Use Linear Fixed Effects Regression Models for Causal Inference with Longitudinal Data?
 When Should We Use Linear Fixed Effects Regression Models for Causal Inference with Longitudinal Data? Kosuke Imai Department of Politics Center for Statistics and Machine Learning Princeton University
When Should We Use Linear Fixed Effects Regression Models for Causal Inference with Longitudinal Data? Kosuke Imai Department of Politics Center for Statistics and Machine Learning Princeton University
INTERPENETRATING SAMPLES
 INTERPENETRATING SAMPLES Dr. A. C. Kulshreshtha UN Statistical Institute for Asia and Pacific Second RAP Regional Workshop on Building Training Resources for Improving Agricultural & Rural Statistics Sampling
INTERPENETRATING SAMPLES Dr. A. C. Kulshreshtha UN Statistical Institute for Asia and Pacific Second RAP Regional Workshop on Building Training Resources for Improving Agricultural & Rural Statistics Sampling
Pooling multiple imputations when the sample happens to be the population.
 Pooling multiple imputations when the sample happens to be the population. Gerko Vink 1,2, and Stef van Buuren 1,3 arxiv:1409.8542v1 [math.st] 30 Sep 2014 1 Department of Methodology and Statistics, Utrecht
Pooling multiple imputations when the sample happens to be the population. Gerko Vink 1,2, and Stef van Buuren 1,3 arxiv:1409.8542v1 [math.st] 30 Sep 2014 1 Department of Methodology and Statistics, Utrecht
Using Phylogenomics to Predict Novel Fungal Pathogenicity Genes
 Using Phylogenomics to Predict Novel Fungal Pathogenicity Genes David DeCaprio, Ying Li, Hung Nguyen (sequenced Ascomycetes genomes courtesy of the Broad Institute) Phylogenomics Combining whole genome
Using Phylogenomics to Predict Novel Fungal Pathogenicity Genes David DeCaprio, Ying Li, Hung Nguyen (sequenced Ascomycetes genomes courtesy of the Broad Institute) Phylogenomics Combining whole genome
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin
 Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
Confidence Intervals in Ridge Regression using Jackknife and Bootstrap Methods
 Chapter 4 Confidence Intervals in Ridge Regression using Jackknife and Bootstrap Methods 4.1 Introduction It is now explicable that ridge regression estimator (here we take ordinary ridge estimator (ORE)
Chapter 4 Confidence Intervals in Ridge Regression using Jackknife and Bootstrap Methods 4.1 Introduction It is now explicable that ridge regression estimator (here we take ordinary ridge estimator (ORE)
8 Error analysis: jackknife & bootstrap
 8 Error analysis: jackknife & bootstrap As discussed before, it is no problem to calculate the expectation values and statistical error estimates of normal observables from Monte Carlo. However, often
8 Error analysis: jackknife & bootstrap As discussed before, it is no problem to calculate the expectation values and statistical error estimates of normal observables from Monte Carlo. However, often
Outline. Genome Evolution. Genome. Genome Architecture. Constraints on Genome Evolution. New Evolutionary Synthesis 11/8/16
 Genome Evolution Outline 1. What: Patterns of Genome Evolution Carol Eunmi Lee Evolution 410 University of Wisconsin 2. Why? Evolution of Genome Complexity and the interaction between Natural Selection
Genome Evolution Outline 1. What: Patterns of Genome Evolution Carol Eunmi Lee Evolution 410 University of Wisconsin 2. Why? Evolution of Genome Complexity and the interaction between Natural Selection
11. Bootstrap Methods
 11. Bootstrap Methods c A. Colin Cameron & Pravin K. Trivedi 2006 These transparencies were prepared in 20043. They can be used as an adjunct to Chapter 11 of our subsequent book Microeconometrics: Methods
11. Bootstrap Methods c A. Colin Cameron & Pravin K. Trivedi 2006 These transparencies were prepared in 20043. They can be used as an adjunct to Chapter 11 of our subsequent book Microeconometrics: Methods
Nonparametric Bayesian Methods (Gaussian Processes)
 [70240413 Statistical Machine Learning, Spring, 2015] Nonparametric Bayesian Methods (Gaussian Processes) Jun Zhu dcszj@mail.tsinghua.edu.cn http://bigml.cs.tsinghua.edu.cn/~jun State Key Lab of Intelligent
[70240413 Statistical Machine Learning, Spring, 2015] Nonparametric Bayesian Methods (Gaussian Processes) Jun Zhu dcszj@mail.tsinghua.edu.cn http://bigml.cs.tsinghua.edu.cn/~jun State Key Lab of Intelligent
On Modifications to Linking Variance Estimators in the Fay-Herriot Model that Induce Robustness
 Statistics and Applications {ISSN 2452-7395 (online)} Volume 16 No. 1, 2018 (New Series), pp 289-303 On Modifications to Linking Variance Estimators in the Fay-Herriot Model that Induce Robustness Snigdhansu
Statistics and Applications {ISSN 2452-7395 (online)} Volume 16 No. 1, 2018 (New Series), pp 289-303 On Modifications to Linking Variance Estimators in the Fay-Herriot Model that Induce Robustness Snigdhansu
Gene Genealogies Coalescence Theory. Annabelle Haudry Glasgow, July 2009
 Gene Genealogies Coalescence Theory Annabelle Haudry Glasgow, July 2009 What could tell a gene genealogy? How much diversity in the population? Has the demographic size of the population changed? How?
Gene Genealogies Coalescence Theory Annabelle Haudry Glasgow, July 2009 What could tell a gene genealogy? How much diversity in the population? Has the demographic size of the population changed? How?
1 Springer. Nan M. Laird Christoph Lange. The Fundamentals of Modern Statistical Genetics
 1 Springer Nan M. Laird Christoph Lange The Fundamentals of Modern Statistical Genetics 1 Introduction to Statistical Genetics and Background in Molecular Genetics 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 Springer Nan M. Laird Christoph Lange The Fundamentals of Modern Statistical Genetics 1 Introduction to Statistical Genetics and Background in Molecular Genetics 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Confidence Estimation Methods for Neural Networks: A Practical Comparison
 , 6-8 000, Confidence Estimation Methods for : A Practical Comparison G. Papadopoulos, P.J. Edwards, A.F. Murray Department of Electronics and Electrical Engineering, University of Edinburgh Abstract.
, 6-8 000, Confidence Estimation Methods for : A Practical Comparison G. Papadopoulos, P.J. Edwards, A.F. Murray Department of Electronics and Electrical Engineering, University of Edinburgh Abstract.
