DIA-Umpire: comprehensive computational framework for data independent acquisition proteomics
|
|
|
- Harvey Fletcher
- 5 years ago
- Views:
Transcription
1 DIA-Umpire: comprehensive computational framework for data independent acquisition proteomics Chih-Chiang Tsou 1,2, Dmitry Avtonomov 2, Brett Larsen 3, Monika Tucholska 3, Hyungwon Choi 4 Anne-Claude Gingras 3,5,*, Alexey I. Nesvizhskii 1,2,* 1. Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan, USA. 2. Department of Pathology, University of Michigan, Ann Arbor, Michigan, USA. 3. Lunenfeld-Tanenbaum Research Institute, Toronto, Canada 4. Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore, Singapore 5. Department of Molecular Genetics, University of Toronto, Toronto, Canada *To whom all correspondence should be addressed: Alexey I. Nesvizhskii nesvi@med.umich.edu; Anne-Claude Gingras gingras@lunenfeld.ca Supplementary Figures and Tables: Supplementary Fig. 1 Example of precursor peptide ion and fragment ion LC elution signals and the corresponding pseudo MS/MS spectrum generated by DIA-Umpire. Supplementary Fig. 2 Effect of MS1 survey scan ion accumulation time on peptide identification using DIA- Umpire. Supplementary Fig. 3 Untargeted peptide identification using DDA and DIA data from human cell lysate samples using three search engines combined. Supplementary Fig. 4 Untargeted peptide identification using DDA and DIA data from E. coli cell lysate samples by X! Tandem search engine. Supplementary Fig. 5 Untargeted peptide identification using DDA and DIA data from E. coli cell lysate samples using three search engines combined. Supplementary Fig. 6 Comparison between untargeted DIA-Umpire analysis and OpenSWATH targeted extraction: effect of the search space. Human cell lysate data. Supplementary Fig. 7 Comparison between untargeted DIA-Umpire analysis and OpenSWATH targeted extraction: effect of the search space. E. coli cell lysate data. Supplementary Fig. 8 Deamidated peptide identifications (the number of peptide ions and unique peptide sequences) from DIA-Umpire and OpenSWATH targeted search. Supplementary Fig. 9 Example of an ambiguous identification involving the deamidated peptide NTTFNVESTK by OpenSWATH targeted search. Supplementary Fig. 10 Example of an ambiguous identification of the deamidated peptide NSPLDEENLTQENQDR by OpenSWATH targeted search. Supplementary Fig. 11 Example of an ambiguous identification of the peptide DIENFNSTQK by OpenSWATH targeted search. 1
2 Supplementary Fig. 12 Example of an ambiguous identification involving the deamidated peptide TGNGLFLSEGLK. Supplementary Fig. 13 Example of an ambiguous identification of the deamidated peptide VAPEEHPTLLTEAPLNPK by OpenSWATH targeted search. Supplementary Fig. 14 Increased identification coverage after targeted re-extraction in DIA-Umpire. Supplementary Fig. 15 MS1-based protein quantification in DIA human cell lysate data. Supplementary Fig. 16 MS2-based protein quantification in DIA human cell lysate data. Supplementary Fig. 17 Comparison between MS1 and MS2-based protein quantification. Supplementary Fig. 18 Protein quantification results in the UPS2 standard protein sample. Supplementary Fig. 19 Distributions of scores computed by the targeted re-extraction algorithm of DIA- Umpire. Supplementary Fig. 20 The numbers of identified proteins and peptide ions via untargeted spectrumcentric search and targeted re-extraction match. Supplementary Fig. 21 Protein quantification in AP-SWATH data. Supplementary Fig. 22 Comparison between DDA and DIA for human cell lysate data generated on a Q Exactive Plus instrument. Supplementary Fig. 23 Visualization of DIA signals using DIA-Umpire result files with Skyline. Supplementary Fig. 24 Illustration of LC-MS data and isotope peaks envelope of a precursor feature. Supplementary Fig. 25 Examples of co-eluting peptide ions. Supplementary Fig. 26. PeptideProphet analysis of X! Tandem search results using DDA and DIA pseudo MS/MS data. Supplementary Fig. 27 Assessment of retention time and MS1 intensity reproducibility of identified peptide ions between DDA and DIA (SWATH) experiments. Human cell lysate data. Supplementary Fig. 28 Assessment of retention time and MS1 intensity reproducibility of identified peptide ions between DDA and DIA (SWATH) experiments. E. coli cell lysate data. Supplementary Fig. 29 Retention time differences for peptide ions commonly identified between DDA replicates. 2
3 Supplementary Fig. 1. Example of precursor peptide ion and fragment ion LC elution signals and the corresponding pseudo MS/MS spectrum generated by DIA-Umpire. (a) Elution profiles for the first 3 isotopic peaks of a doubly charged precursor peptide ion AMGIM[Oxy]NSFVNDIFER extracted from MS1 data from a DIA (SWATH) run on a AB SCIEX 5600 instrument. (b) Elution profiles for fragments of this precursor peptide detected in the DIA MS2 data. (c) DDA MS/MS spectrum (from a DDA run generated on the same instrument and using the same sample) from which the same peptide was identified, with matched b- and y- ions highlighted. (d) Pseudo MS/MS spectrum extracted by DIA-Umpire from the DIA data (before complementary ion boosting). (e) Same pseudo MS/MS spectrum after complementary ion boosting. Note a larger number of b- ions matched in (e) compared to (d). (a) and (b) images exported from Skyline. (c), (d), and (e) are exported from TPP spectrum browser. 3
4 Supplementary Fig. 2 Effect of MS1 survey scan ion accumulation time on peptide identification using DIA-Umpire. Experiments to assess the identification performance of DIA-Umpire on data generated using different MS1 ion accumulation times in DIA (SWATH) analysis using AB SCIEX 5600 instrument were carried out using two samples: UPS1 proteins, and UPS2 mixture spiked in with E. coli background. Two settings (50 ms and 250 ms MS1 ion accumulation time) were tested. Three replicate runs were acquired for each sample/condition. The numbers shown are non-redundant contributions to the total number of peptide ion identifications in each replicate/condition from pseudo MS/MS spectra from three different quality tiers: QT = 1 (white bar), 2 (grey), and 3 (dark grey). The QT = 1 category represents pseudo MS/MS spectra that are linked to high quality MS1 precursor features (3 or more detected isotope peaks), QT = 2 represent lower abundance precursors (2 detected isotope peaks only), and QT = 3 represents unfragmented precursors detected in DIA MS2 scans. In a low complexity UPS1 sample, the dominant majority of peptide ions were identified from QT =1 spectra. Even with the using short MS1 accumulation time (50 ms), 92 93% of the peptides ions were identified from the QT = 1 spectral subset (this fraction increased slightly, to 94 96%, with the longer 250 ms accumulation time). Note that inclusion of unfragmented precursors detected in DIA MS2 data (QT = 3 subset) in the analysis contributed 4 6% of the total peptide ion identifications in UPS1 samples. In the more complex UPS2 plus E. coli samples, the effect of the accumulation time on the quality of MS1 signal was more pronounced. The longer DIA MS1 survey scan ion accumulation time resulted in more high quality (QT = 1) precursor peptide features detected, and thus more peptides identified from pseudo MS/MS spectra in the QT = 1 subset (81 85% for 250 ms vs % for 50 ms). Congruently, QT = 2 and QT = 3 spectral subsets contributed a higher percentage to the total number of peptide ion identifications when using 50 ms accumulation time setting. The overall number of identifications (from all 3 QT sets) has improved with 250 ms vs. 50 ms acquisition time (~ 10%). Overall, this analysis indicates that longer MS1 accumulation time provides an advantage to DIA-Umpire algorithm with respect to the total number of identified peptide ions, especially peptide ions identified with a high quality MS1 precursor ion signal. 4
5 Supplementary Fig. 3. Untargeted peptide identification using DDA and DIA data from human cell lysate samples using three search engines combined. DIA pseudo MS/MS spectra were searched using X! Tandem, Comet, and MSGF+, and combined using iprophet. Protein and peptide ion identifications were then filtered at 1 % FDR using target-decoy approach. (a) The numbers of proteins and peptide ions identified at 1% FDR in DDA and in DIA data. Left: number of protein identifications in each experiment (1,831 proteins identified from DDA data, 1,692 from DIA, 1,964 in total). Right: Total number of peptide ion identifications from two replicates (10,822 peptide ions identified from DDA data, 10,922 from DIA, 14,997 in total). Compared to using X! Tandem only (main text, Figure 4) when the results from all three search 5
6 engines were combined the number of identifications increased in both DDA (by 17% and 11% for peptide ions and proteins, respectively) and in DIA data (by 25% and 16% for peptide ions and proteins, respectively). However, the overlap between the DIA and DDA identified peptide ions and proteins increased only slightly, to 45% and 79%. Of the peptide ions identified by DIA and not DDA at 1% FDR (total 4,175 peptide ions), the majority of the remaining peptide ions were not identified by DDA because no MS/MS was acquired (2,742). (b) Percent of fragments ions matched in pseudo MS/MS spectra extracted from DIA data as a function of the MS1 peptide ion identity in DDA data. Data points (peptide ions) and the summary density plots are labeled according to the three categories of peptide ions: ions identified from DIA data at 1% FDR ( Identified in DIA ; blue), and unidentified in DIA (orange; these ions were located in DIA data as described in Online Methods). (c) Comparison between DDA and DIA in terms of numbers of fragments matched among two categories of peptide ions, showing that peptide ions identified with confidence from DDA but not DIA have fewer fragment ions that could be matched. 6
7 Supplementary Fig. 4. Untargeted peptide identification using DDA and DIA data from E. coli cell lysate samples with X! Tandem search engine. Results for E. coli data were similar to those obtained for human cell lysate data (see Fig. 4). 7
8 Supplementary Fig. 5. Untargeted peptide identification using DDA and DIA data from E. coli cell lysate samples with three search engines combined. Results for E. coli data were similar to those obtained for human cell lysate when using X! Tandem, Comet, and MSGF+ (combined using iprophet; see Supplementary Fig. 3). 8
9 Supplementary Fig. 6 Comparison between untargeted DIA-Umpire analysis and OpenSWATH targeted extraction: effect of the search space. Human cell lysate data (a) The pseudo MS/MS spectra extracted using DIA-Umpire were searched against two sequence databases: Whole proteome contains all proteins in the human proteome (plus decoy proteins); Library peptide database contains only the sequences of the DDA identified peptides (i.e. it is built using the same peptides as the spectral library used 9
10 for targeted extraction with OpenSWATH). The total numbers of candidate peptide ions considered for scoring of pseudo MS/MS spectra extracted by DIA-Umpire during database search against Whole proteome or Library peptide databases were estimated using the following parameters: 30 ppm precursor mass tolerance; peptide sequence length: 4 ~ 50 amino acids; one missed cleavage allowed; charge state considered: 2+, 3+, or 4+; m/z range: 350 ~ 1200 Da; variable modifications: oxidation of methionine, cysteine alkylation, conversion of pyroglutamate from glutamine or glutamic acid, and n-terminal acetylation (allowing less than six modifications on the same peptide). For OpenSWATH analysis, the following parameters were used to estimate number of candidate fragment groups in the experimental SWATH MS2 data considered for each target library peptide: mass range of the corresponding SWATH m/z isolation (25 Da wide) and ± 1 minute retention time window. The use of the precursor ion m/z value from MS1 or MS2 unfragmented precursors as a constraint during database search was the primary factor contributing to the significant reduction in the number of candidate peptide ions considered for scoring against each spectrum (from 68,344,142 peptides in the whole proteome database to 4,960 searched ions per spectrum on average, i.e. 13,779 fold reduction). Because targeted data extraction in OpenSWATH used the retention time of the peptide and wide (25 Da) m/z SWATH selection window (but not the precursor peptide m/z) for constraining the search space, the reduction in the number of candidates was less significant (from 18,544 peptide ions in the library to 31 ions per ± 1 minute retention time slice of the corresponding 25 Da MS2 SWATH scan, i.e. 598 fold reduction). The order of magnitude difference in the search space reduction in DIA-Umpire/ Whole proteome analysis (compared to OpenSWATH/Library analysis) explains why DIA- Umpire untargeted analysis performed well. (b) Venn diagram of peptide ion identifications among the three analyses. DIA-Umpire/Whole proteome and OpenSWATH/Library identified a comparable number of peptide ions, but the two methods had only a moderate overlap. OpenSWATH identified larger fraction of peptides in the library, 79% (i.e. (4, ,458 = 7,372) / 9,272) vs. 58% (i.e. ( = 5,369) / 9,272)) for DIA-Umpire. At the same time, DIA-Umpire was able to identify a large number of peptide ions not present in the library. DIA-Umpire/ Library peptide analysis had an effective search space similar to that of OpenSWATH, resulting in even closer performance (and better overlap) between the two methods: the overlap between the DIA-Umpire identified peptides and the DDA-identified peptides improved to 69% (or (5, ) / 9,272 peptide ions) (c) Venn diagram of protein identifications among the three analyses (Whole proteome sequence database was used for DIA-Umpire). 10
11 Supplementary Fig. 7 Comparison between untargeted DIA-Umpire analysis and OpenSWATH targeted extraction: effect of the search space. E. coli cell lysate data. Results similar to those presented from human cell lysate were obtained for E. coli data (see Supplementary Fig. 6) 11
12 Supplementary Fig. 8 Deamidated peptide identifications (the number of peptide ions and unique peptide sequences) from DIA-Umpire and OpenSWATH targeted search. Glycoproteomics data from Liu et al. Mol Cell Proteomics (2014). Upper panel: all peptides. Lower panel: peptides containing the NX(S/T) motif expected to be significantly enriched in these data. 12
13 Supplementary Fig. 9 Example of an ambiguous identification involving the deamidated peptide NTTFNVESTK by OpenSWATH targeted search. Although retention times of different modified/unmodified peptide species can help resolve the ambiguity, how different modifications influence retention time remains an open problem for computational prediction. If both species exist in the library and only one of them is present in the sample, library spectra from both species might match the same fragment peak groups in the queried DIA MS2 data. Although the correct one should get a higher matching score, the score of the incorrect one is likely to be better than any decoy peptide and thus also deemed a confident identification. In the glycoproteomics application presented in this work, identification of N-linked glycopeptides depends on detection of asparagine deamidation in peptides due to PNGase F treatment, which causes only a small mass shift (0.984 Da), resulting in both modified and unmodified peptides being co-fragmented. Therefore, we searched the OpenSWATH data for identifications of both deamidated and non-deamidated species reported as highly confident (m_score < 0.01), at the same retention time (within 1 second), and of the same charge state. Here we present one example (see Supplementary Figs for additional examples) in which OpenSWATH was not able to distinguish deamidated peptide ions from unmodified peptide ions (we manually checked these by using the exact precursor mass). More specifically, two separate identifications with different modification site compositions (with one and two deamidations; modification site shown in red) were reported by OpenSWATH. The two identifications both had an extremely small m_score (from mprophet), i.e. they both were reported as high confidence identifications. The two identifications had identical retention times. The MS1 signal image shown above suggests there is only one peptide eluting at RT = 36.9 minutes (precursor m/z of Da). DIA-Umpire reported only one (singly deamidated) form, further supported by the presence of NXS/T motif covering the reported site. This example demonstrates that the knowledge of the precursor mass can be very valuable for differentiating between different modification forms of the same peptide sequence in DIA experiments. 13
14 Supplementary Fig. 10 Example of an ambiguous identification of the deamidated peptide NSPLDEENLTQENQDR by OpenSWATH targeted search. Two separate identifications (in unmodified form and in a deamidated form; the site of the modification is shown in red) were reported by OpenSWATH. The two identifications both had an extremely small m_score (from mprophet), i.e. they both were reported as high confidence identifications. The two identifications had identical retention times. The MS1 signal image shown above suggests there is only one peptide eluting at RT = minutes (precursor m/z of Da). DIA-Umpire reported only one (unmodified) form. 14
15 Supplementary Fig. 11 Example of an ambiguous identification of the peptide DIENFNSTQK by OpenSWATH targeted search. Two separate identifications with different modification site compositions (with one and two deamidations; modification site shown in red) were reported by OpenSWATH. The two identifications both had a small m_score (from mprophet), i.e. they both were reported as high confidence identifications. The two identifications had almost identical retention times (within 0.02 minute). The MS1 signal image shown above suggests there is only one peptide eluting at RT = 37.4 minutes (precursor m/z of Da). DIA-Umpire reported only one (singly deamidated) form, further supported by the presence of NXS/T motif covering the reported site. 15
16 Supplementary Fig. 12 Example of an ambiguous identification involving the deamidated peptide TGNGLFLSEGLK. Two separate identifications were reported (in unmodified and in deamidated form) by OpenSWATH. The two identifications both had an extremely small m_score (from mprophet), i.e. they both were reported as high confidence identifications. The two identifications had identical retention times. The MS1 signal image shown above suggests there is only one peptide eluting at RT = 67.6 minutes (precursor m/z of Da), which was identified by DIA-Umpire as unmodified peptide. In addition, DIA-Umpire identified the deamidated form of the peptide at retention time of minutes (also marked on the MS1 signal image). 16
17 Supplementary Fig. 13 Example of an ambiguous identification of the deamidated peptide VAPEEHPTLLTEAPLNPK by OpenSWATH targeted search. Two separate identifications (in unmodified form and in a deamidated form; the site of the modification is shown in red) were reported by OpenSWATH. The two identifications both had an extremely small m_score (from mprophet), i.e. they both were reported as high confidence identifications. The two identifications had almost identical retention times. The MS1 signal image shown above suggests there is only one peptide eluting at RT = minutes (precursor m/z of Da). DIA-Umpire reported only one (unmodified) form. 17
18 Supplementary Fig. 14 Increased identification coverage after targeted re-extraction in DIA-Umpire. Human cell lysate DIA data. (a) Venn diagram of peptide ion identifications in two replicates of human cell lysate DIA (SWATH) data from untargeted X! Tandem search; (b) the number of peptide ions identified in both replicates increased after targeted re-extraction; (c) same as (a) at the protein level; (d) same as (b) at the protein level. 18
19 Supplementary Fig. 15 MS1-based protein quantification in DIA human cell lysate data. (a) MS1 ibaq intensity; (b) MS1 Top3pep (indep. selection) : protein intensity estimated by summing the top three most intense peptide ions, independently in each DIA run; (c) MS1 Top3pep, Freq>0.5 : same as (b), with an additional requirement that the selected peptides are identified in more than 50% of the runs (Freq > 0.5) in which the corresponding protein was identified; (d) MS1 Top6pep, Freq>0.5 : same as (c), but using six most intense peptides. Note that selection of consistently identified peptide ions (Freq > 0.5 filter) significantly improves the reproducibility of protein intensities between the replicates. 19
20 Supplementary Fig. 16 MS2-based protein quantification in DIA human cell lysate data. (a) MS2 ibaq intensity; (b) MS2 Top3pep/Top2fra (indep. selection) : protein intensity estimated by summing the top three most intense peptide ions. Peptide ion intensities are estimated by summing the intensities of their top 2 most intense matched fragments. Protein intensities are computed independently for each DIA run; (c) MS2 Top3pep/Top2fra : same as (b), but using peptide ions and fragments having the highest overall intensity across all runs (here, highest summed intensity across the two replicates); (d) same as (c), but using six selected peptide ions and fragments, with an additional requirement that the selected peptides and fragments are identified in more than 50% of the runs (Freq > 0.5) in which the corresponding protein (peptide selection step) or peptide (fragment selection step) was identified. Note that selection of consistently identified peptide ions and fragments (Freq > 0.5 filter) significantly improves the reproducibility of protein intensities between the replicates. 20
21 Supplementary Fig. 17 Comparison between MS1 and MS2-based protein quantification. Human cell lysate data. MS1-based intensities are computed using the Top6pep, Freq>0.5 method. MS2-based protein intensities are computed using the Top6pep/Top6fra, Freq>0.5 method. 21
22 Supplementary Fig. 18 Protein quantification results in the UPS2 standard protein sample. In UPS2 samples, protein concentrations are known and span five orders of magnitude (8 proteins in each of the five abundance bins; no proteins were quantified in the lowest abundance bin in this study). Proteins were quantified using four quantification methods. (a) MS1 ibaq intensity; (b) MS2 ibaq intensity; (c) MS1 Top6pep, Freq>0.5 : protein intensity estimated by summing the top six most intense peptide ions which are consistently identified (Freq > 0.5 filter); (d) MS2 Top6pep/Top6fra, Freq>0.5 : protein intensity estimated by summing the top six most intense peptide ions with an additional requirement that the selected peptides and fragments are identified in more than 50% of the runs (Freq > 0.5) in which the corresponding protein (peptide selection step) or peptide (fragment selection step) was identified. 22
23 Supplementary Fig. 19 Distributions of scores computed by the targeted re-extraction algorithm of DIA-Umpire. AP-SWATH dataset, MEPCE bait (biological replicate 3). (a) Distributions of the five subscores for peptide ions from the positive set ( Identified features, i.e. precursor-fragment group to spectrum matches that were confidently identified by the untargeted spectrum-centric search) and from the negative set (precursor-fragment groups matching to decoy spectra); (b) Linear discriminant analysis is used to train the weights in the linear combination used to combine the individual sub-scores to compute a single discriminant score (U-score). Shown are the resulting U-score distributions for positive and negative matches in the training set. (c) The final distribution of U-scores for all non-decoy matches. The observed distribution is model as a mixture of two underlying distribution representing high scoring, correct matches (red curve) and low scoring, incorrect matches (blue curve). The parameters of the distributions are learned using the expectation maximization mixture modeling algorithm. The posterior probability of a correct match is computed for a given non-decoy spectrum to precursor-fragments group match from the ratio of learned distributions among correct and incorrect matches. By default, peptide ions with a computed probability above 0.99 are considered confidently identified and contribute, together with the peptide ions identified at the initial untargeted identification stage, to protein quantification for their corresponding protein. 23
24 Supplementary Fig. 20 The numbers of identified proteins and peptide ions via untargeted spectrum-centric search and targeted re-extraction matching. The numbers of identified peptide ions and proteins from the initial untargeted (spectrum-centric search) analysis using DIA-Umpire (blue bars) shown separately for 3 biological replicates (Biorep1 Biorep3) of the two bait proteins (EIF4A2 and MEPCE) and the GFP negative controls. Also shown (red bars) the numbers of additional peptide ions and proteins identified by the targeted re-extraction using the spectral library internally generated from the initial search results. 24
25 Supplementary Fig. 21 Protein quantification in AP-SWATH data. Protein intensities are computed using the MS2 Top6pep/Top6fra, Freq>0.5 approach. Each dot represents computed protein intensities for the same protein in two different biological replicates for the same bait (or GFP control). 25
26 Supplementary Fig. 22 Comparison between DDA and DIA for human cell lysate data generated on a Q Exactive Plus instrument. The samples were prepared in the same way as the samples that were analyzed using the AB SCIEX 5600 instrument used to generate the main datasets used in the manuscript. Peptide samples were analyzed using an EasySpray column (25cm x 75um x 2um C18) with 90 minute gradient at 300nl/min coupled online to a Q Exactive Plus (QE) instrument. (a) Number of identified peptide ions between QE DDA and QE DIA. (b) Detailed analysis for fragment loss and MS1 intensity for different 26
27 categories of peptide ions: high scoring DIA (blue), low scoring DIA (red), and unidentified in DIA (orange). (c) A detailed comparison between DDA and DIA in terms of the numbers of fragments matched among three categories of peptide ions. 27
28 Supplementary Fig. 23 Visualization of DIA signals using DIA-Umpire result files with Skyline. 28
29 Supplementary Fig. 24 Illustration of LC-MS data and isotope peaks envelope of a precursor feature. Images exported using OpenMS
30 Supplementary Fig. 25 Examples of co-eluting peptide ions. (a) Two co-eluted peptide ions A and B with the monoisotopic peak A 1 of peptide ion A overlapped with the third isotope peak B 3 of peptide ion B. The peak detection algorithms have a difficulty with detecting B 3 because it is completely buried by A 1 signal. (b) Another, more complicated example where co-elution of multiple peptide ions presents an ambiguity with the interpretation of different isotope peak groups. To effectively detect as many true precursor ions as possible, the signal detection algorithm of DIA-Umpire considers each peak curve as a possible monoisotopic peak, and then attempts to find higher isotope peak curves for the assumed monoisotopic peak. 30
31 Supplementary Fig. 26. PeptideProphet analysis of X! Tandem search results using DDA and DIA pseudo MS/MS data. Shown are model distributions learned by PeptideProphet in the analysis of X! Tandem search results (doubly charged peptide ions) for one replicate of the human cell lysate data. Left panels: mass accuracy distributions. Right panels: the distributions of the discriminant database search scores (computed from the X! Tandem expect scores). Red and blue curves represent the models learned by PeptideProphet for correct and incorrect identifications, respectively. Also shown are the distributions for the number of missed cleavages parameter (nmc) among correct and incorrect identifications. (a) DDA data; (b) DIA data, QT=1 pseudo MS/MS spectra; (c) DIA data, QT=2 spectra. The learned distributions appear to be an accurate fit in both DIA and DDA data, demonstrating that the search results obtained using DIA pseudo MS/MS spectra can be satisfactory analyzed using PeptideProphet. The overall higher ratio of incorrect vs correct identification in the DIA QT=1 vs. DDA data (and similarly in DIA QT=2 vs. QT=1 data) simply reflects the higher number of pseudo MS/MS spectra extracted from the data compared to DDA data (and similarly, more noise in DIA QT=2 vs QT=1 data), which does not affect the accuracy of computed PeptideProphet probabilities or the subsequent FDR estimates. 31
32 Supplementary Fig. 27 Assessment of retention time and MS1 intensity reproducibility of identified peptide ions between DDA and DIA (SWATH) experiments. Human cell lysate data. (a) LC retention time (LC peak apex) for peptide ions identified in both DDA and DIA experiments. (b) MS1 intensities (monoisotopic peak intensities at LC peak apex) of peptide ions identified commonly by DDA and DIA. (c) Reproducibility of DIA MS1 peptide ion intensities between two replicates of DIA data. (d) Reproducibility of DIA MS2 fragment ion intensities (at the reconstructed LC peak apex) of peptide ions between two DIA replicates. Only matched b- and y-ion fragments were considered. Ion and fragment intensities are shown on log 2 scale. 32
33 Supplementary Fig. 28 Assessment of retention time and MS1 intensity reproducibility of identified peptide ions between DDA and DIA (SWATH) experiments. E. coli cell lysate data. (a) LC retention time (LC peak apex) for peptide ions identified in both DDA and DIA experiments. (b) MS1 intensities (monoisotopic peak intensities at LC peak apex) of peptide ions identified commonly by DDA and DIA. (c) Reproducibility of DIA MS1 peptide ion intensities between two replicates of DIA data. (d) Reproducibility of DIA MS2 fragment ion intensities (at the reconstructed LC peak apex) of peptide ions between two DIA replicates. Only matched b- and y-ion fragments were considered. Ion and fragment intensities are shown on log 2 scale. 33
34 Supplementary Fig. 29 Retention time differences for peptide ions commonly identified between DDA replicates. (a) E. coli cell lysate data (b) human cell lysate data. 34
Nature Methods: doi: /nmeth Supplementary Figure 1. Fragment indexing allows efficient spectra similarity comparisons.
 Supplementary Figure 1 Fragment indexing allows efficient spectra similarity comparisons. The cost and efficiency of spectra similarity calculations can be approximated by the number of fragment comparisons
Supplementary Figure 1 Fragment indexing allows efficient spectra similarity comparisons. The cost and efficiency of spectra similarity calculations can be approximated by the number of fragment comparisons
Tutorial 2: Analysis of DIA data in Skyline
 Tutorial 2: Analysis of DIA data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantitation using
Tutorial 2: Analysis of DIA data in Skyline In this tutorial we will learn how to use Skyline to perform targeted post-acquisition analysis for peptide and inferred protein detection and quantitation using
Tutorial 1: Setting up your Skyline document
 Tutorial 1: Setting up your Skyline document Caution! For using Skyline the number formats of your computer have to be set to English (United States). Open the Control Panel Clock, Language, and Region
Tutorial 1: Setting up your Skyline document Caution! For using Skyline the number formats of your computer have to be set to English (United States). Open the Control Panel Clock, Language, and Region
Tutorial 1: Library Generation from DDA data
 Tutorial 1: Library Generation from DDA data 1. Introduction Before a targeted, peptide-centric DIA analysis can be performed, a spectral library containing peptide-query parameters needs to be generated.
Tutorial 1: Library Generation from DDA data 1. Introduction Before a targeted, peptide-centric DIA analysis can be performed, a spectral library containing peptide-query parameters needs to be generated.
TUTORIAL EXERCISES WITH ANSWERS
 TUTORIAL EXERCISES WITH ANSWERS Tutorial 1 Settings 1. What is the exact monoisotopic mass difference for peptides carrying a 13 C (and NO additional 15 N) labelled C-terminal lysine residue? a. 6.020129
TUTORIAL EXERCISES WITH ANSWERS Tutorial 1 Settings 1. What is the exact monoisotopic mass difference for peptides carrying a 13 C (and NO additional 15 N) labelled C-terminal lysine residue? a. 6.020129
SRM assay generation and data analysis in Skyline
 in Skyline Preparation 1. Download the example data from www.srmcourse.ch/eupa.html (3 raw files, 1 csv file, 1 sptxt file). 2. The number formats of your computer have to be set to English (United States).
in Skyline Preparation 1. Download the example data from www.srmcourse.ch/eupa.html (3 raw files, 1 csv file, 1 sptxt file). 2. The number formats of your computer have to be set to English (United States).
Protein Quantitation II: Multiple Reaction Monitoring. Kelly Ruggles New York University
 Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot RPPA Immunohistochemistry
Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot RPPA Immunohistochemistry
Improved 6- Plex TMT Quantification Throughput Using a Linear Ion Trap HCD MS 3 Scan Jane M. Liu, 1,2 * Michael J. Sweredoski, 2 Sonja Hess 2 *
 Improved 6- Plex TMT Quantification Throughput Using a Linear Ion Trap HCD MS 3 Scan Jane M. Liu, 1,2 * Michael J. Sweredoski, 2 Sonja Hess 2 * 1 Department of Chemistry, Pomona College, Claremont, California
Improved 6- Plex TMT Quantification Throughput Using a Linear Ion Trap HCD MS 3 Scan Jane M. Liu, 1,2 * Michael J. Sweredoski, 2 Sonja Hess 2 * 1 Department of Chemistry, Pomona College, Claremont, California
Protein Quantitation II: Multiple Reaction Monitoring. Kelly Ruggles New York University
 Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot Immunohistochemistry
Protein Quantitation II: Multiple Reaction Monitoring Kelly Ruggles kelly@fenyolab.org New York University Traditional Affinity-based proteomics Use antibodies to quantify proteins Western Blot Immunohistochemistry
HOWTO, example workflow and data files. (Version )
 HOWTO, example workflow and data files. (Version 20 09 2017) 1 Introduction: SugarQb is a collection of software tools (Nodes) which enable the automated identification of intact glycopeptides from HCD
HOWTO, example workflow and data files. (Version 20 09 2017) 1 Introduction: SugarQb is a collection of software tools (Nodes) which enable the automated identification of intact glycopeptides from HCD
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra. Andrew Keller
 PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Outline Need to validate peptide assignments to MS/MS spectra Statistical approach to validation Running PeptideProphet
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Outline Need to validate peptide assignments to MS/MS spectra Statistical approach to validation Running PeptideProphet
MassHunter TOF/QTOF Users Meeting
 MassHunter TOF/QTOF Users Meeting 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks
MassHunter TOF/QTOF Users Meeting 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks
Protein Identification Using Tandem Mass Spectrometry. Nathan Edwards Informatics Research Applied Biosystems
 Protein Identification Using Tandem Mass Spectrometry Nathan Edwards Informatics Research Applied Biosystems Outline Proteomics context Tandem mass spectrometry Peptide fragmentation Peptide identification
Protein Identification Using Tandem Mass Spectrometry Nathan Edwards Informatics Research Applied Biosystems Outline Proteomics context Tandem mass spectrometry Peptide fragmentation Peptide identification
Overview - MS Proteomics in One Slide. MS masses of peptides. MS/MS fragments of a peptide. Results! Match to sequence database
 Overview - MS Proteomics in One Slide Obtain protein Digest into peptides Acquire spectra in mass spectrometer MS masses of peptides MS/MS fragments of a peptide Results! Match to sequence database 2 But
Overview - MS Proteomics in One Slide Obtain protein Digest into peptides Acquire spectra in mass spectrometer MS masses of peptides MS/MS fragments of a peptide Results! Match to sequence database 2 But
Analysis of Labeled and Non-Labeled Proteomic Data Using Progenesis QI for Proteomics
 Analysis of Labeled and Non-Labeled Proteomic Data Using Progenesis QI for Proteomics Lee Gethings, Gushinder Atwal, Martin Palmer, Chris Hughes, Hans Vissers, and James Langridge Waters Corporation, Wilmslow,
Analysis of Labeled and Non-Labeled Proteomic Data Using Progenesis QI for Proteomics Lee Gethings, Gushinder Atwal, Martin Palmer, Chris Hughes, Hans Vissers, and James Langridge Waters Corporation, Wilmslow,
MassHunter Software Overview
 MassHunter Software Overview 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks A
MassHunter Software Overview 1 Qualitative Analysis Workflows Workflows in Qualitative Analysis allow the user to only see and work with the areas and dialog boxes they need for their specific tasks A
Spectrum-to-Spectrum Searching Using a. Proteome-wide Spectral Library
 MCP Papers in Press. Published on April 30, 2011 as Manuscript M111.007666 Spectrum-to-Spectrum Searching Using a Proteome-wide Spectral Library Chia-Yu Yen, Stephane Houel, Natalie G. Ahn, and William
MCP Papers in Press. Published on April 30, 2011 as Manuscript M111.007666 Spectrum-to-Spectrum Searching Using a Proteome-wide Spectral Library Chia-Yu Yen, Stephane Houel, Natalie G. Ahn, and William
Designed for Accuracy. Innovation with Integrity. High resolution quantitative proteomics LC-MS
 Designed for Accuracy High resolution quantitative proteomics Innovation with Integrity LC-MS Setting New Standards in Accuracy The development of mass spectrometry based proteomics approaches has dramatically
Designed for Accuracy High resolution quantitative proteomics Innovation with Integrity LC-MS Setting New Standards in Accuracy The development of mass spectrometry based proteomics approaches has dramatically
Mass Spectrometry and Proteomics - Lecture 5 - Matthias Trost Newcastle University
 Mass Spectrometry and Proteomics - Lecture 5 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Proteomics Sample prep 144 Lecture 5 Quantitation techniques Search Algorithms Proteomics
Mass Spectrometry and Proteomics - Lecture 5 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Proteomics Sample prep 144 Lecture 5 Quantitation techniques Search Algorithms Proteomics
Targeted protein quantification
 Targeted Quantitative Proteomics Targeted protein quantification with high-resolution, accurate-mass MS Highly selective Very sensitive Complex samples HR/AM A more complete quantitative proteomics picture
Targeted Quantitative Proteomics Targeted protein quantification with high-resolution, accurate-mass MS Highly selective Very sensitive Complex samples HR/AM A more complete quantitative proteomics picture
MS-MS Analysis Programs
 MS-MS Analysis Programs Basic Process Genome - Gives AA sequences of proteins Use this to predict spectra Compare data to prediction Determine degree of correctness Make assignment Did we see the protein?
MS-MS Analysis Programs Basic Process Genome - Gives AA sequences of proteins Use this to predict spectra Compare data to prediction Determine degree of correctness Make assignment Did we see the protein?
Computational Methods for Mass Spectrometry Proteomics
 Computational Methods for Mass Spectrometry Proteomics Eidhammer, Ingvar ISBN-13: 9780470512975 Table of Contents Preface. Acknowledgements. 1 Protein, Proteome, and Proteomics. 1.1 Primary goals for studying
Computational Methods for Mass Spectrometry Proteomics Eidhammer, Ingvar ISBN-13: 9780470512975 Table of Contents Preface. Acknowledgements. 1 Protein, Proteome, and Proteomics. 1.1 Primary goals for studying
Modeling Mass Spectrometry-Based Protein Analysis
 Chapter 8 Jan Eriksson and David Fenyö Abstract The success of mass spectrometry based proteomics depends on efficient methods for data analysis. These methods require a detailed understanding of the information
Chapter 8 Jan Eriksson and David Fenyö Abstract The success of mass spectrometry based proteomics depends on efficient methods for data analysis. These methods require a detailed understanding of the information
iprophet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates
 MCP Papers in Press. Published on August 29, 2011 as Manuscript M111.007690 This is the Pre-Published Version iprophet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein
MCP Papers in Press. Published on August 29, 2011 as Manuscript M111.007690 This is the Pre-Published Version iprophet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein
Proteomics. November 13, 2007
 Proteomics November 13, 2007 Acknowledgement Slides presented here have been borrowed from presentations by : Dr. Mark A. Knepper (LKEM, NHLBI, NIH) Dr. Nathan Edwards (Center for Bioinformatics and Computational
Proteomics November 13, 2007 Acknowledgement Slides presented here have been borrowed from presentations by : Dr. Mark A. Knepper (LKEM, NHLBI, NIH) Dr. Nathan Edwards (Center for Bioinformatics and Computational
All Ions MS/MS: Targeted Screening and Quantitation Using Agilent TOF and Q-TOF LC/MS Systems
 All Ions MS/MS: Targeted Screening and Quantitation Using Agilent TOF and Q-TOF LC/MS Systems Technical Overview Introduction All Ions MS/MS is a technique that is available for Agilent high resolution
All Ions MS/MS: Targeted Screening and Quantitation Using Agilent TOF and Q-TOF LC/MS Systems Technical Overview Introduction All Ions MS/MS is a technique that is available for Agilent high resolution
WADA Technical Document TD2015IDCR
 MINIMUM CRITERIA FOR CHROMATOGRAPHIC-MASS SPECTROMETRIC CONFIRMATION OF THE IDENTITY OF ANALYTES FOR DOPING CONTROL PURPOSES. The ability of a method to identify an analyte is a function of the entire
MINIMUM CRITERIA FOR CHROMATOGRAPHIC-MASS SPECTROMETRIC CONFIRMATION OF THE IDENTITY OF ANALYTES FOR DOPING CONTROL PURPOSES. The ability of a method to identify an analyte is a function of the entire
Workflow concept. Data goes through the workflow. A Node contains an operation An edge represents data flow The results are brought together in tables
 PROTEOME DISCOVERER Workflow concept Data goes through the workflow Spectra Peptides Quantitation A Node contains an operation An edge represents data flow The results are brought together in tables Protein
PROTEOME DISCOVERER Workflow concept Data goes through the workflow Spectra Peptides Quantitation A Node contains an operation An edge represents data flow The results are brought together in tables Protein
Targeted Proteomics Environment
 Targeted Proteomics Environment Quantitative Proteomics with Bruker Q-TOF Instruments and Skyline Brendan MacLean Quantitative Proteomics Spectrum-based Spectral counting Isobaric tags Chromatography-based
Targeted Proteomics Environment Quantitative Proteomics with Bruker Q-TOF Instruments and Skyline Brendan MacLean Quantitative Proteomics Spectrum-based Spectral counting Isobaric tags Chromatography-based
WADA Technical Document TD2003IDCR
 IDENTIFICATION CRITERIA FOR QUALITATIVE ASSAYS INCORPORATING CHROMATOGRAPHY AND MASS SPECTROMETRY The appropriate analytical characteristics must be documented for a particular assay. The Laboratory must
IDENTIFICATION CRITERIA FOR QUALITATIVE ASSAYS INCORPORATING CHROMATOGRAPHY AND MASS SPECTROMETRY The appropriate analytical characteristics must be documented for a particular assay. The Laboratory must
Received: December 16, 2016 Accepted: April 9, 2017 Published: April 9, 2017
 This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/ac Downloaded
This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/ac Downloaded
Multi-residue analysis of pesticides by GC-HRMS
 An Executive Summary Multi-residue analysis of pesticides by GC-HRMS Dr. Hans Mol is senior scientist at RIKILT- Wageningen UR Introduction Regulatory authorities throughout the world set and enforce strict
An Executive Summary Multi-residue analysis of pesticides by GC-HRMS Dr. Hans Mol is senior scientist at RIKILT- Wageningen UR Introduction Regulatory authorities throughout the world set and enforce strict
Self-assembling covalent organic frameworks functionalized. magnetic graphene hydrophilic biocomposite as an ultrasensitive
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2017 Electronic Supporting Information for: Self-assembling covalent organic frameworks functionalized
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2017 Electronic Supporting Information for: Self-assembling covalent organic frameworks functionalized
Feature File 1. Feature Detection. Similarity Comparison (Precursor and product ions) Spectra Generation. Decoy spectra.
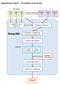 Supplementary Figure 1. The workflow of Group-DIA. DIA Data Data File 1 Data File 2 Data File 3 Feature Detection Feature File 1 Feature File 2 Feature File 3 Retention Time Alignment Intensity Extraction
Supplementary Figure 1. The workflow of Group-DIA. DIA Data Data File 1 Data File 2 Data File 3 Feature Detection Feature File 1 Feature File 2 Feature File 3 Retention Time Alignment Intensity Extraction
6 x 5 Ways to Ensure Your LC-MS/MS is Healthy
 6 x 5 Ways to Ensure Your LC-MS/MS is Healthy (Also known as - Tracking Performance with the 6 x 5 LC-MS/MS Peptide Reference Mixture) Mike Rosenblatt, Ph.D. Group Leader Mass Spec Reagents 215. We monitor
6 x 5 Ways to Ensure Your LC-MS/MS is Healthy (Also known as - Tracking Performance with the 6 x 5 LC-MS/MS Peptide Reference Mixture) Mike Rosenblatt, Ph.D. Group Leader Mass Spec Reagents 215. We monitor
Analyst Software. Peptide and Protein Quantitation Tutorial
 This document is provided to customers who have purchased AB Sciex equipment to use in the operation of such AB Sciex equipment. This document is copyright protected and any reproduction of this document
This document is provided to customers who have purchased AB Sciex equipment to use in the operation of such AB Sciex equipment. This document is copyright protected and any reproduction of this document
Identification of a Set of Conserved Eukaryotic Internal Retention Time Standards for Data-Independent Acquisition Mass Spectrometry
 MCP Papers in Press. Published on July 21, 2015 as Manuscript O114.042267 Identification of a Set of Conserved Eukaryotic Internal Retention Time Standards for Data-Independent Acquisition Mass Spectrometry
MCP Papers in Press. Published on July 21, 2015 as Manuscript O114.042267 Identification of a Set of Conserved Eukaryotic Internal Retention Time Standards for Data-Independent Acquisition Mass Spectrometry
Model-based Biclustering of Label-free Quantitative AP-MS Data"
 1 Supplementary Document for "Analysis of Protein Complexes via Model-based Biclustering of Label-free Quantitative AP-MS Data" Table of Contents Search Parameters for PP2A Dataset... Page 2 Prior Elicitation
1 Supplementary Document for "Analysis of Protein Complexes via Model-based Biclustering of Label-free Quantitative AP-MS Data" Table of Contents Search Parameters for PP2A Dataset... Page 2 Prior Elicitation
Supplementary Figure 1
 Supplementary Figure 1 The correlation of n-score cutoff and FDR in both CID-only and CID-ETD fragmentation strategies. A bar diagram of different n-score thresholds applied in the search, plotted against
Supplementary Figure 1 The correlation of n-score cutoff and FDR in both CID-only and CID-ETD fragmentation strategies. A bar diagram of different n-score thresholds applied in the search, plotted against
Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis
 Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis Technical Overview Introduction Metabolomics studies measure the relative abundance of metabolites
Agilent MassHunter Profinder: Solving the Challenge of Isotopologue Extraction for Qualitative Flux Analysis Technical Overview Introduction Metabolomics studies measure the relative abundance of metabolites
Skyline Small Molecule Targets
 Skyline Small Molecule Targets The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline documents. Originally developed
Skyline Small Molecule Targets The Skyline Targeted Proteomics Environment provides informative visual displays of the raw mass spectrometer data you import into your Skyline documents. Originally developed
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra
 PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Day 2 October 17, 2006 Andrew Keller Rosetta Bioinformatics, Seattle Outline Need to validate peptide assignments to MS/MS
PeptideProphet: Validation of Peptide Assignments to MS/MS Spectra Andrew Keller Day 2 October 17, 2006 Andrew Keller Rosetta Bioinformatics, Seattle Outline Need to validate peptide assignments to MS/MS
Figure S1. Interaction of PcTS with αsyn. (a) 1 H- 15 N HSQC NMR spectra of 100 µm αsyn in the absence (0:1, black) and increasing equivalent
 Figure S1. Interaction of PcTS with αsyn. (a) 1 H- 15 N HSQC NMR spectra of 100 µm αsyn in the absence (0:1, black) and increasing equivalent concentrations of PcTS (100 µm, blue; 500 µm, green; 1.5 mm,
Figure S1. Interaction of PcTS with αsyn. (a) 1 H- 15 N HSQC NMR spectra of 100 µm αsyn in the absence (0:1, black) and increasing equivalent concentrations of PcTS (100 µm, blue; 500 µm, green; 1.5 mm,
Methods for proteome analysis of obesity (Adipose tissue)
 Methods for proteome analysis of obesity (Adipose tissue) I. Sample preparation and liquid chromatography-tandem mass spectrometric analysis Instruments, softwares, and materials AB SCIEX Triple TOF 5600
Methods for proteome analysis of obesity (Adipose tissue) I. Sample preparation and liquid chromatography-tandem mass spectrometric analysis Instruments, softwares, and materials AB SCIEX Triple TOF 5600
Isotopic-Labeling and Mass Spectrometry-Based Quantitative Proteomics
 Isotopic-Labeling and Mass Spectrometry-Based Quantitative Proteomics Xiao-jun Li, Ph.D. Current address: Homestead Clinical Day 4 October 19, 2006 Protein Quantification LC-MS/MS Data XLink mzxml file
Isotopic-Labeling and Mass Spectrometry-Based Quantitative Proteomics Xiao-jun Li, Ph.D. Current address: Homestead Clinical Day 4 October 19, 2006 Protein Quantification LC-MS/MS Data XLink mzxml file
Comprehensive support for quantitation
 Comprehensive support for quantitation One of the major new features in the current release of Mascot is support for quantitation. This is still work in progress. Our goal is to support all of the popular
Comprehensive support for quantitation One of the major new features in the current release of Mascot is support for quantitation. This is still work in progress. Our goal is to support all of the popular
The Pitfalls of Peaklist Generation Software Performance on Database Searches
 Proceedings of the 56th ASMS Conference on Mass Spectrometry and Allied Topics, Denver, CO, June 1-5, 2008 The Pitfalls of Peaklist Generation Software Performance on Database Searches Aenoch J. Lynn,
Proceedings of the 56th ASMS Conference on Mass Spectrometry and Allied Topics, Denver, CO, June 1-5, 2008 The Pitfalls of Peaklist Generation Software Performance on Database Searches Aenoch J. Lynn,
TOMAHAQ Method Construction
 TOMAHAQ Method Construction Triggered by offset mass accurate-mass high-resolution accurate quantitation (TOMAHAQ) can be performed in the standard method editor of the instrument, without modifications
TOMAHAQ Method Construction Triggered by offset mass accurate-mass high-resolution accurate quantitation (TOMAHAQ) can be performed in the standard method editor of the instrument, without modifications
Metabolomics in an Identity Crisis? Am I a Feature or a Compound? The world leader in serving science
 Metabolomics in an Identity Crisis? Am I a Feature or a Compound? The world leader in serving science Agenda 1 2 3 Unknown Analysis in Metabolomics Determining Features and Compounds for Data Reduction
Metabolomics in an Identity Crisis? Am I a Feature or a Compound? The world leader in serving science Agenda 1 2 3 Unknown Analysis in Metabolomics Determining Features and Compounds for Data Reduction
Site-specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells
 Supplementary Information Site-specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells Tong Zhou 1, 2, Ying-hua Chung 1, 2, Jianji Chen 1, Yue Chen 1 1. Department of Biochemistry,
Supplementary Information Site-specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells Tong Zhou 1, 2, Ying-hua Chung 1, 2, Jianji Chen 1, Yue Chen 1 1. Department of Biochemistry,
Tandem mass spectra were extracted from the Xcalibur data system format. (.RAW) and charge state assignment was performed using in house software
 Supplementary Methods Software Interpretation of Tandem mass spectra Tandem mass spectra were extracted from the Xcalibur data system format (.RAW) and charge state assignment was performed using in house
Supplementary Methods Software Interpretation of Tandem mass spectra Tandem mass spectra were extracted from the Xcalibur data system format (.RAW) and charge state assignment was performed using in house
Relative quantification using TMT11plex on a modified Q Exactive HF mass spectrometer
 POSTER NOTE 6558 Relative quantification using TMT11plex on a modified mass spectrometer Authors Tabiwang N. Arrey, 1 Rosa Viner, 2 Ryan D. Bomgarden, 3 Eugen Damoc, 1 Markus Kellmann, 1 Thomas Moehring,
POSTER NOTE 6558 Relative quantification using TMT11plex on a modified mass spectrometer Authors Tabiwang N. Arrey, 1 Rosa Viner, 2 Ryan D. Bomgarden, 3 Eugen Damoc, 1 Markus Kellmann, 1 Thomas Moehring,
Proteome-wide label-free quantification with MaxQuant. Jürgen Cox Max Planck Institute of Biochemistry July 2011
 Proteome-wide label-free quantification with MaxQuant Jürgen Cox Max Planck Institute of Biochemistry July 2011 MaxQuant MaxQuant Feature detection Data acquisition Initial Andromeda search Statistics
Proteome-wide label-free quantification with MaxQuant Jürgen Cox Max Planck Institute of Biochemistry July 2011 MaxQuant MaxQuant Feature detection Data acquisition Initial Andromeda search Statistics
Spectronaut Pulsar. User Manual
 Spectronaut Pulsar User Manual 1 General Information... 6 1.1 Computer System Requirements... 6 1.2 Scope of Spectronaut Software... 6 1.3 Spectronaut Pulsar... 6 1.4 Spectronaut Release Features... 7
Spectronaut Pulsar User Manual 1 General Information... 6 1.1 Computer System Requirements... 6 1.2 Scope of Spectronaut Software... 6 1.3 Spectronaut Pulsar... 6 1.4 Spectronaut Release Features... 7
A TMT-labeled Spectral Library for Peptide Sequencing
 A TMT-labeled Spectral Library for Peptide Sequencing by Jianqiao Shen A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree of Master of Mathematics
A TMT-labeled Spectral Library for Peptide Sequencing by Jianqiao Shen A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree of Master of Mathematics
High-Field Orbitrap Creating new possibilities
 Thermo Scientific Orbitrap Elite Hybrid Mass Spectrometer High-Field Orbitrap Creating new possibilities Ultrahigh resolution Faster scanning Higher sensitivity Complementary fragmentation The highest
Thermo Scientific Orbitrap Elite Hybrid Mass Spectrometer High-Field Orbitrap Creating new possibilities Ultrahigh resolution Faster scanning Higher sensitivity Complementary fragmentation The highest
QTOF-based proteomics and metabolomics for the agro-food chain.
 QTOF-based proteomics and metabolomics for the agro-food chain luigi.lucini@unicatt.it Metabolomics Two scenarios identification of known unknowns and unknown unknowns For known unknowns use spectral or
QTOF-based proteomics and metabolomics for the agro-food chain luigi.lucini@unicatt.it Metabolomics Two scenarios identification of known unknowns and unknown unknowns For known unknowns use spectral or
An extended siderophore suite from Synechococcus sp. PCC 7002 revealed by LC- ICPMS-ESIMS
 Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 2015 An extended siderophore suite from Synechococcus sp. PCC 7002 revealed by LC- ICPMS-ESIMS Rene
Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 2015 An extended siderophore suite from Synechococcus sp. PCC 7002 revealed by LC- ICPMS-ESIMS Rene
Key questions of proteomics. Bioinformatics 2. Proteomics. Foundation of proteomics. What proteins are there? Protein digestion
 s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture 2 roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture 2 roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
NEW TOOLS FOR FINDING AND IDENTIFYING METABOLITES IN A METABOLOMICS WORKFLOW
 NEW TOOLS FOR FINDING AND IDENTIFYING METABOLITES IN A METABOLOMICS WORKFLOW Julia E. Wingate 1 ; Elliott Jones 2 ; Armin Graber 3 ; Klaus Weinberger 3 1Applied Biosystems, Toronto, Canada; 2Applied Biosystems,
NEW TOOLS FOR FINDING AND IDENTIFYING METABOLITES IN A METABOLOMICS WORKFLOW Julia E. Wingate 1 ; Elliott Jones 2 ; Armin Graber 3 ; Klaus Weinberger 3 1Applied Biosystems, Toronto, Canada; 2Applied Biosystems,
Effective Strategies for Improving Peptide Identification with Tandem Mass Spectrometry
 Effective Strategies for Improving Peptide Identification with Tandem Mass Spectrometry by Xi Han A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree
Effective Strategies for Improving Peptide Identification with Tandem Mass Spectrometry by Xi Han A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree
An Effective Workflow for Impurity Analysis Incorporating High Quality HRAM LCMS & MSMS with Intelligent Automated Data Mining
 An Effective Workflow for Impurity Analysis Incorporating High Quality HRAM LCMS & MSMS with Intelligent Automated Data Mining Dave Weil, Ph.D. and Jim Lau, Ph.D. Typical Method Conditions: 1260 UHPLC
An Effective Workflow for Impurity Analysis Incorporating High Quality HRAM LCMS & MSMS with Intelligent Automated Data Mining Dave Weil, Ph.D. and Jim Lau, Ph.D. Typical Method Conditions: 1260 UHPLC
Last updated: Copyright
 Last updated: 2012-08-20 Copyright 2004-2012 plabel (v2.4) User s Manual by Bioinformatics Group, Institute of Computing Technology, Chinese Academy of Sciences Tel: 86-10-62601016 Email: zhangkun01@ict.ac.cn,
Last updated: 2012-08-20 Copyright 2004-2012 plabel (v2.4) User s Manual by Bioinformatics Group, Institute of Computing Technology, Chinese Academy of Sciences Tel: 86-10-62601016 Email: zhangkun01@ict.ac.cn,
NPTEL VIDEO COURSE PROTEOMICS PROF. SANJEEVA SRIVASTAVA
 LECTURE-25 Quantitative proteomics: itraq and TMT TRANSCRIPT Welcome to the proteomics course. Today we will talk about quantitative proteomics and discuss about itraq and TMT techniques. The quantitative
LECTURE-25 Quantitative proteomics: itraq and TMT TRANSCRIPT Welcome to the proteomics course. Today we will talk about quantitative proteomics and discuss about itraq and TMT techniques. The quantitative
Mass Spectrometry Based De Novo Peptide Sequencing Error Correction
 Mass Spectrometry Based De Novo Peptide Sequencing Error Correction by Chenyu Yao A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree of Master of Mathematics
Mass Spectrometry Based De Novo Peptide Sequencing Error Correction by Chenyu Yao A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree of Master of Mathematics
Key Words Q Exactive, Accela, MetQuest, Mass Frontier, Drug Discovery
 Metabolite Stability Screening and Hotspot Metabolite Identification by Combining High-Resolution, Accurate-Mass Nonselective and Selective Fragmentation Tim Stratton, Caroline Ding, Yingying Huang, Dan
Metabolite Stability Screening and Hotspot Metabolite Identification by Combining High-Resolution, Accurate-Mass Nonselective and Selective Fragmentation Tim Stratton, Caroline Ding, Yingying Huang, Dan
Improved Validation of Peptide MS/MS Assignments. Using Spectral Intensity Prediction
 MCP Papers in Press. Published on October 2, 2006 as Manuscript M600320-MCP200 Improved Validation of Peptide MS/MS Assignments Using Spectral Intensity Prediction Shaojun Sun 1, Karen Meyer-Arendt 2,
MCP Papers in Press. Published on October 2, 2006 as Manuscript M600320-MCP200 Improved Validation of Peptide MS/MS Assignments Using Spectral Intensity Prediction Shaojun Sun 1, Karen Meyer-Arendt 2,
PC235: 2008 Lecture 5: Quantitation. Arnold Falick
 PC235: 2008 Lecture 5: Quantitation Arnold Falick falickam@berkeley.edu Summary What you will learn from this lecture: There are many methods to perform quantitation using mass spectrometry (any method
PC235: 2008 Lecture 5: Quantitation Arnold Falick falickam@berkeley.edu Summary What you will learn from this lecture: There are many methods to perform quantitation using mass spectrometry (any method
STATISTICAL METHODS FOR THE ANALYSIS OF MASS SPECTROMETRY- BASED PROTEOMICS DATA. A Dissertation XUAN WANG
 STATISTICAL METHODS FOR THE ANALYSIS OF MASS SPECTROMETRY- BASED PROTEOMICS DATA A Dissertation by XUAN WANG Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of
STATISTICAL METHODS FOR THE ANALYSIS OF MASS SPECTROMETRY- BASED PROTEOMICS DATA A Dissertation by XUAN WANG Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of
Yifei Bao. Beatrix. Manor Askenazi
 Detection and Correction of Interference in MS1 Quantitation of Peptides Using their Isotope Distributions Yifei Bao Department of Computer Science Stevens Institute of Technology Beatrix Ueberheide Department
Detection and Correction of Interference in MS1 Quantitation of Peptides Using their Isotope Distributions Yifei Bao Department of Computer Science Stevens Institute of Technology Beatrix Ueberheide Department
Increasing Speed of UHPLC-MS Analysis Using Single-stage Orbitrap Mass Spectrometer
 Increasing Speed of UHPLC-MS Analysis Using Single-stage Orbitrap Mass Spectrometer Olaf Scheibner and Maciej Bromirski Thermo Fisher Scientific, Bremen, Germany Overview Purpose: Improve the performance
Increasing Speed of UHPLC-MS Analysis Using Single-stage Orbitrap Mass Spectrometer Olaf Scheibner and Maciej Bromirski Thermo Fisher Scientific, Bremen, Germany Overview Purpose: Improve the performance
Compounding insights Thermo Scientific Compound Discoverer Software
 Compounding insights Thermo Scientific Compound Discoverer Software Integrated, complete, toolset solves small-molecule analysis challenges Thermo Scientific Orbitrap mass spectrometers produce information-rich
Compounding insights Thermo Scientific Compound Discoverer Software Integrated, complete, toolset solves small-molecule analysis challenges Thermo Scientific Orbitrap mass spectrometers produce information-rich
Quantitation of a target protein in crude samples using targeted peptide quantification by Mass Spectrometry
 Quantitation of a target protein in crude samples using targeted peptide quantification by Mass Spectrometry Jon Hao, Rong Ye, and Mason Tao Poochon Scientific, Frederick, Maryland 21701 Abstract Background:
Quantitation of a target protein in crude samples using targeted peptide quantification by Mass Spectrometry Jon Hao, Rong Ye, and Mason Tao Poochon Scientific, Frederick, Maryland 21701 Abstract Background:
A Workflow Approach for the Identification and Structural Elucidation of Impurities of Quetiapine Hemifumarate Drug Substance
 A Workflow Approach for the Identification and Structural Elucidation of Impurities of Quetiapine Hemifumarate Drug Substance Michael D. Jones, Marian Twohig, Karen Haas, and Robert S. Plumb Waters Corporation,
A Workflow Approach for the Identification and Structural Elucidation of Impurities of Quetiapine Hemifumarate Drug Substance Michael D. Jones, Marian Twohig, Karen Haas, and Robert S. Plumb Waters Corporation,
HR/AM Targeted Peptide Quantification on a Q Exactive MS: A Unique Combination of High Selectivity, High Sensitivity, and High Throughput
 HR/AM Targeted Peptide Quantification on a Q Exactive MS: A Unique Combination of High Selectivity, High Sensitivity, and High Throughput Yi Zhang 1, Zhiqi Hao 1, Markus Kellmann 2 and Andreas FR. Huhmer
HR/AM Targeted Peptide Quantification on a Q Exactive MS: A Unique Combination of High Selectivity, High Sensitivity, and High Throughput Yi Zhang 1, Zhiqi Hao 1, Markus Kellmann 2 and Andreas FR. Huhmer
MS-based proteomics to investigate proteins and their modifications
 MS-based proteomics to investigate proteins and their modifications Francis Impens VIB Proteomics Core October th 217 Overview Mass spectrometry-based proteomics: general workflow Identification of protein
MS-based proteomics to investigate proteins and their modifications Francis Impens VIB Proteomics Core October th 217 Overview Mass spectrometry-based proteomics: general workflow Identification of protein
Overview. Introduction. André Schreiber AB SCIEX Concord, Ontario (Canada)
 Quantitation and Identification of Pharmaceuticals and Personal Care Products (PPCP) in Environmental Samples using Advanced TripleTOF MS/MS Technology André Schreiber AB SCIEX Concord, Ontario (Canada)
Quantitation and Identification of Pharmaceuticals and Personal Care Products (PPCP) in Environmental Samples using Advanced TripleTOF MS/MS Technology André Schreiber AB SCIEX Concord, Ontario (Canada)
High-Throughput Protein Quantitation Using Multiple Reaction Monitoring
 High-Throughput Protein Quantitation Using Multiple Reaction Monitoring Application Note Authors Ning Tang, Christine Miller, Joe Roark, Norton Kitagawa and Keith Waddell Agilent Technologies, Inc. Santa
High-Throughput Protein Quantitation Using Multiple Reaction Monitoring Application Note Authors Ning Tang, Christine Miller, Joe Roark, Norton Kitagawa and Keith Waddell Agilent Technologies, Inc. Santa
Mass spectrometry has been used a lot in biology since the late 1950 s. However it really came into play in the late 1980 s once methods were
 Mass spectrometry has been used a lot in biology since the late 1950 s. However it really came into play in the late 1980 s once methods were developed to allow the analysis of large intact (bigger than
Mass spectrometry has been used a lot in biology since the late 1950 s. However it really came into play in the late 1980 s once methods were developed to allow the analysis of large intact (bigger than
Learning Score Function Parameters for Improved Spectrum Identification in Tandem Mass Spectrometry Experiments
 pubs.acs.org/jpr Learning Score Function Parameters for Improved Spectrum Identification in Tandem Mass Spectrometry Experiments Marina Spivak, Michael S. Bereman, Michael J. MacCoss, and William Stafford
pubs.acs.org/jpr Learning Score Function Parameters for Improved Spectrum Identification in Tandem Mass Spectrometry Experiments Marina Spivak, Michael S. Bereman, Michael J. MacCoss, and William Stafford
Analysis of Peptide MS/MS Spectra from Large-Scale Proteomics Experiments Using Spectrum Libraries
 Anal. Chem. 2006, 78, 5678-5684 Analysis of Peptide MS/MS Spectra from Large-Scale Proteomics Experiments Using Spectrum Libraries Barbara E. Frewen, Gennifer E. Merrihew, Christine C. Wu, William Stafford
Anal. Chem. 2006, 78, 5678-5684 Analysis of Peptide MS/MS Spectra from Large-Scale Proteomics Experiments Using Spectrum Libraries Barbara E. Frewen, Gennifer E. Merrihew, Christine C. Wu, William Stafford
The Power of LC MALDI: Identification of Proteins by LC MALDI MS/MS Using the Applied Biosystems 4700 Proteomics Analyzer with TOF/TOF Optics
 APPLICATION NOTE TOF MS The Power of LC MALDI: Identification of Proteins by LC MALDI MS/MS Using the Applied Biosystems 4700 Proteomics Analyzer with TOF/TOF Optics Purpose The Applied Biosystems 4700
APPLICATION NOTE TOF MS The Power of LC MALDI: Identification of Proteins by LC MALDI MS/MS Using the Applied Biosystems 4700 Proteomics Analyzer with TOF/TOF Optics Purpose The Applied Biosystems 4700
Peter A. DiMaggio, Jr., Nicolas L. Young, Richard C. Baliban, Benjamin A. Garcia, and Christodoulos A. Floudas. Research
 Research A Mixed Integer Linear Optimization Framework for the Identification and Quantification of Targeted Post-translational Modifications of Highly Modified Proteins Using Multiplexed Electron Transfer
Research A Mixed Integer Linear Optimization Framework for the Identification and Quantification of Targeted Post-translational Modifications of Highly Modified Proteins Using Multiplexed Electron Transfer
Tandem MS = MS / MS. ESI-MS give information on the mass of a molecule but none on the structure
 Tandem MS = MS / MS ESI-MS give information on the mass of a molecule but none on the structure In tandem MS (MSMS) (pseudo-)molecular ions are selected in MS1 and fragmented by collision with gas. collision
Tandem MS = MS / MS ESI-MS give information on the mass of a molecule but none on the structure In tandem MS (MSMS) (pseudo-)molecular ions are selected in MS1 and fragmented by collision with gas. collision
Relative Quantitation of TMT-Labeled Proteomes Focus on Sensitivity and Precision
 Relative Quantitation of TMT-Labeled Proteomes Focus on Sensitivity and Precision R. Viner 1, M. Scigelova 2, M. Zeller 2, M. Oppermann 2, T. Moehring 2 and V. Zabrouskov 1 1 Thermo Fisher Scientific,
Relative Quantitation of TMT-Labeled Proteomes Focus on Sensitivity and Precision R. Viner 1, M. Scigelova 2, M. Zeller 2, M. Oppermann 2, T. Moehring 2 and V. Zabrouskov 1 1 Thermo Fisher Scientific,
PRIDE Cluster: building the consensus of proteomics data
 Supplementary Materials PRIDE Cluster: building the consensus of proteomics data Johannes Griss, Joseph Michael Foster, Henning Hermjakob and Juan Antonio Vizcaíno EMBL-European Bioinformatics Institute,
Supplementary Materials PRIDE Cluster: building the consensus of proteomics data Johannes Griss, Joseph Michael Foster, Henning Hermjakob and Juan Antonio Vizcaíno EMBL-European Bioinformatics Institute,
Development of a protein quantification algorithm for data analysis in the field of proteomics
 Facultat d Informàtica de Barcelona (FIB) Development of a protein quantification algorithm for data analysis in the field of proteomics Autora: Ariadna Llovet Soto Titulació: Enginyeria superior en informàtica
Facultat d Informàtica de Barcelona (FIB) Development of a protein quantification algorithm for data analysis in the field of proteomics Autora: Ariadna Llovet Soto Titulació: Enginyeria superior en informàtica
Chemical Labeling Strategy for Generation of Internal Standards for Targeted Quantitative Proteomics
 Chemical Labeling Strategy for Generation of Internal Standards for Targeted Quantitative Proteomics mtraq Reagents Triplex Christie Hunter, Brian Williamson, Marjorie Minkoff AB SCIEX, USA The utility
Chemical Labeling Strategy for Generation of Internal Standards for Targeted Quantitative Proteomics mtraq Reagents Triplex Christie Hunter, Brian Williamson, Marjorie Minkoff AB SCIEX, USA The utility
De Novo Peptide Sequencing: Informatics and Pattern Recognition applied to Proteomics
 De Novo Peptide Sequencing: Informatics and Pattern Recognition applied to Proteomics John R. Rose Computer Science and Engineering University of South Carolina 1 Overview Background Information Theoretic
De Novo Peptide Sequencing: Informatics and Pattern Recognition applied to Proteomics John R. Rose Computer Science and Engineering University of South Carolina 1 Overview Background Information Theoretic
X!TandemPipeline (Myosine Anabolisée) validating, filtering and grouping MSMS identifications
 X!TandemPipeline 3.3.3 (Myosine Anabolisée) validating, filtering and grouping MSMS identifications Olivier Langella and Benoit Valot langella@moulon.inra.fr; valot@moulon.inra.fr PAPPSO - http://pappso.inra.fr/
X!TandemPipeline 3.3.3 (Myosine Anabolisée) validating, filtering and grouping MSMS identifications Olivier Langella and Benoit Valot langella@moulon.inra.fr; valot@moulon.inra.fr PAPPSO - http://pappso.inra.fr/
FUTURE CONFIRMATORY CRITERIA
 FUTURE CONFIRMATORY CRITERIA Revision CD 2002/657 Marco Blokland, Bjorn Berendsen, Hans Mol, Leen van Ginkel, Saskia Sterk REVISION 2 Revision 2002/657 EU Commission has explicitly asked for a revision
FUTURE CONFIRMATORY CRITERIA Revision CD 2002/657 Marco Blokland, Bjorn Berendsen, Hans Mol, Leen van Ginkel, Saskia Sterk REVISION 2 Revision 2002/657 EU Commission has explicitly asked for a revision
De novo Protein Sequencing by Combining Top-Down and Bottom-Up Tandem Mass Spectra. Xiaowen Liu
 De novo Protein Sequencing by Combining Top-Down and Bottom-Up Tandem Mass Spectra Xiaowen Liu Department of BioHealth Informatics, Department of Computer and Information Sciences, Indiana University-Purdue
De novo Protein Sequencing by Combining Top-Down and Bottom-Up Tandem Mass Spectra Xiaowen Liu Department of BioHealth Informatics, Department of Computer and Information Sciences, Indiana University-Purdue
MetWorks Metabolite Identification Software
 m a s s s p e c t r o m e t r y MetWorks Metabolite Identification Software Enabling Confident Analysis of Metabolism Data Part of Thermo Fisher Scientific MetWorks Software for the Confident Analysis
m a s s s p e c t r o m e t r y MetWorks Metabolite Identification Software Enabling Confident Analysis of Metabolism Data Part of Thermo Fisher Scientific MetWorks Software for the Confident Analysis
Tandem Mass Spectrometry: Generating function, alignment and assembly
 Tandem Mass Spectrometry: Generating function, alignment and assembly With slides from Sangtae Kim and from Jones & Pevzner 2004 Determining reliability of identifications Can we use Target/Decoy to estimate
Tandem Mass Spectrometry: Generating function, alignment and assembly With slides from Sangtae Kim and from Jones & Pevzner 2004 Determining reliability of identifications Can we use Target/Decoy to estimate
Reagents. Affinity Tag (Biotin) Acid Cleavage Site. Figure 1. Cleavable ICAT Reagent Structure.
 DATA SHEET Protein Expression Analysis Reagents Background The ultimate goal of proteomics is to identify and quantify proteins that are relevant to a given biological state; and to unearth networks of
DATA SHEET Protein Expression Analysis Reagents Background The ultimate goal of proteomics is to identify and quantify proteins that are relevant to a given biological state; and to unearth networks of
MSblender: a probabilistic approach for integrating peptide identifications from multiple database search engines
 Article Subscriber access provided by University of Texas Libraries MSblender: a probabilistic approach for integrating peptide identifications from multiple database search engines Taejoon Kwon, Hyungwon
Article Subscriber access provided by University of Texas Libraries MSblender: a probabilistic approach for integrating peptide identifications from multiple database search engines Taejoon Kwon, Hyungwon
SPECTRA LIBRARY ASSISTED DE NOVO PEPTIDE SEQUENCING FOR HCD AND ETD SPECTRA PAIRS
 SPECTRA LIBRARY ASSISTED DE NOVO PEPTIDE SEQUENCING FOR HCD AND ETD SPECTRA PAIRS 1 Yan Yan Department of Computer Science University of Western Ontario, Canada OUTLINE Background Tandem mass spectrometry
SPECTRA LIBRARY ASSISTED DE NOVO PEPTIDE SEQUENCING FOR HCD AND ETD SPECTRA PAIRS 1 Yan Yan Department of Computer Science University of Western Ontario, Canada OUTLINE Background Tandem mass spectrometry
Increasing the Multiplexing of Protein Quantitation from 6- to 10-Plex with Reporter Ion Isotopologues
 Increasing the Multiplexing of Protein Quantitation from 6- to 1-Plex with Reporter Ion Isotopologues Rosa Viner, 1 Ryan Bomgarden, 2 Michael Blank, 1 John Rogers 2 1 Thermo Fisher Scientific, San Jose,
Increasing the Multiplexing of Protein Quantitation from 6- to 1-Plex with Reporter Ion Isotopologues Rosa Viner, 1 Ryan Bomgarden, 2 Michael Blank, 1 John Rogers 2 1 Thermo Fisher Scientific, San Jose,
Agilent MassHunter Quantitative Data Analysis
 Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch Table, Compound Information Setup, Calibration Curve and Globals Settings 1 MassHunter
Agilent MassHunter Quantitative Data Analysis Presenters: Howard Sanford Stephen Harnos MassHunter Quantitation: Batch Table, Compound Information Setup, Calibration Curve and Globals Settings 1 MassHunter
Developing Algorithms for the Determination of Relative Abundances of Peptides from LC/MS Data
 Developing Algorithms for the Determination of Relative Abundances of Peptides from LC/MS Data RIPS Team Jake Marcus (Project Manager) Anne Eaton Melanie Kanter Aru Ray Faculty Mentors Shawn Cokus Matteo
Developing Algorithms for the Determination of Relative Abundances of Peptides from LC/MS Data RIPS Team Jake Marcus (Project Manager) Anne Eaton Melanie Kanter Aru Ray Faculty Mentors Shawn Cokus Matteo
