Molegro Molecular Viewer (MMV) Program de vizualizare off-line a proteinelor si a interactiunii ligand - proteina
|
|
|
- Heather Banks
- 6 years ago
- Views:
Transcription
1 Molegro Molecular Viewer (MMV) Program de vizualizare off-line a proteinelor si a interactiunii ligand - proteina
2 Molegro Molecular Viewer - aplicatie pentru studiul si analiza interactiunii dintre liganzi si proteine (macromolecule). Programul permite deschiderea fisierelor cu extensia pdb, salvate din baza de date RCSB Protein Data Bank (rcsb.org/pdb).
3 Fisierele proteinei de interes se descarca de pe site-ul (rcsb.org), din pe pagina dedicate proteinei respective. Pentru deschiderea fisierului unei proteine se foloseste comanda import
4 MMV se bazeaza pe notiunea de "workspaces". Un "spatiu de lucru" poate fi salvat, curatat, inlocuit sau lipit la alte "spatii de lucru".
5 Molegro Molecular Viewer toolbar: - usureaza accesul la actiunile uzuale de vizualizare a unor molecule. Import molecule Fit to screen Screen capture Search Label dialog Visualization settings Hydrogens On/Off
6 Fereastra Workspace Explorer: contine informatii despre: - molecule (proteine, liganzi, moleculele de apa) - etichete (labels), - suprafete, - scheletul proteinei (backbones), - cavitati Butonul dreapta al mouse-ului (context menu) permite: Exportul moleculei (format PDB, Mol2, SDF) Editare in fereastra Workspace Properties Redenumirea moleculei Eliminarea unor elemente din fereastra activa Clonarea ligandului sau a proteinei (copy) Convertirea proteinei la ligand Prepararea de molecule Crearea de etichete, suprafete si schelete (backbones) Fitarea moleculei in Visualization Window
7 Fereastra Workspace Explorer poate fi folosita pentru a inspecta molecula in Visualization Window (click pe molecula de interes)
8 Properties Window contine informatii despre obiectele selectate in Visualization Window
9 Visualization Window: pentru vizualizarea de proteine, liganzi, suprafete, cavitati, molecule de apa (doar oxigen), etichete, notatii, scheletul proteinei, legaturi de hidrogen,... Modificarea modului de afisare se face activand Visualization settings : - stilul si culoarea moleculelor, - rendering options, - legaturile de hydrogen, - interactiunile electrostatice
10 Style and color : moleculele pot fi reprezentate ca - ball-and-sticks, - sticks, - space-fill (CPK), - wireframe Ball and Stick : atomii sunt sfere iar legaturile sunt reprezentate ca cilindrii. Atom Scale seteaza fractia razei Van der Waals care este folosita ca raza a sferelor. Bond Scale este diametrul legaturii in Angstrom Reprezentarea Ball-and-stick este folosita pentru prepararea liganzilor: legatura vizualizata indica ordinul legaturii, culoarea ne spune daca legatura este rigida (brown or red) sau flexibila (green). Stick : legaturile sunt figurate ca cilindrii. Bond Scale este diametrul legaturii in Angstrom Spacefill (CPK). Atomii sunt figurati ca sfere. Legaturile nu sunt vizualizate Atom Scale seteaza fractia razei Van der Waals care este folosita ca raza a sferelor. Wireframe cel mai rapid mod de desenare a moleculelor: legaturile sunt fogurate ca linii intre atomi. Atomii nu sunt figurati (dar este posibil sa fie selectati). Toate legaturile sunt desenate cu o singura linie indifferent daca sunt duble, simple sau delocalizate.
11 Setari coloring styles ce pot fi aplicate tuturor moleculelor Fixed Color Color By Element (CPK) Color By Id (or Chain) Color By Id (carbons only) Color By Hydrogen Bond Type (rosu = donori, verde = acceptori, galben = atomi capabili si sa doneze si sa accepte atomi de hidrogen). Color By Partial Charge (albastru = pozitiv, rosu = negativ) Setari coloring styles ce pot fi aplicate numai proteinelor Color By Temperature (B-Factor) (albastru = Tmin, rosu = Tmax) factorul B (Temperatura) indica masura in care atomul vibreaza in jurul pozitiei de echilibru Color By Amino Acid Type Color By Shapely Residue Color By Residue ID (rainbow effect). Color By Secondary Structure (rosu = -helices, albastru = -strands, galben = -turns. Color By Hydrophobicity (rosu = aminoacid hydrophilic, albastru = amoinoacid hydrophobic.
12 Rendering: modificarea setarilor de afisaj: fog, 3D projection, background, label Interactions: hydrogen bonds; electrostatic interactions Views: moduri presetate de reprezentare
13 Toate rotatiile se fac in jurul unui centru de rotatie care poate fi ales: Set as Rotational Center Toate obiectele din Visualization Window (spatiul 3D) pot fi actionate prin "context menu" (click dreapta)
14 Console Window (in josul ecranului) afiseaza informatiii, avertizari si erori
15 Se pot crea vizualizari ale suprafetei moleculei: Tools ; Surfaces. Se selecteaza molecula dorita (proteina, ligand, etc) Se selecteaza tipul de suprafata (electrostatic, hidrofobicitate, )
16 Clipping Planes: modificarea planului de vizualizare (ex. pentru vizualizarea interiorului unei proteine) Se activeaza din meniul Window : Se ajusteaza planul apropiat si cel indepartat pana cand este vizualizata regiunea dorita.
17 Search Space: poate fi folosit pentru a crea o suprafata moleculara partiala: - se ascund moleculele in exteriorul spatiului creat; Se seteaza centrul suprafetei: - centrul proteinei - centrul ligandului (daca exista) Se seteaza raza sferei
18 Hide Residues permite ascunderea reziduurilor (aminoacizilor) din exteriorul unei sfere predefinite (centru, raza) Crop molecules: stergerea unor parti din molecula (eliminarea regiunilor nerelevante). Workspace Finder: cautarea rapida a reziduurilor/atomilor sau a moleculelor.
19 Sequence Viewer: (se activeaza din "Tools" sau "Window"): permite inspectarea rezidurilor proteinei. Pentru evidentiere se selecteaza aminoacidul Pentru informatii se plaseaza cursorul pe aminoacidul respectiv
20 Masuratori si notatii: Daca se selecteaza 2 atomi: - in Properties Window este afisata distanta dintre ei. Daca se selecteaza 3 atomi conectati: - in Properties Window este afisat unghiul dintre ei. Daca se selecteaza o legatura: - in Properties Window este afisat unghiul de torsiune definit prin aceasta legatura. Pentru a face diferite notatii: - se selecteaza 1 atom "click dreapta Create Point Annotation - se selecteaza 2 atomi "click dreapta Create Distance Annotation - se selecteaza 3 atomi "click dreapta Create Angle Annotation - se selecteaza 4 atomi "click dreapta Create Dihedral Angle Annotation
21 - Create Backbone: din "context menu" (click dreapta) pe categoria "Protein" in Workspace Explorer - cartoon style (vizualizarea structurii secundare) - tube style (scheletul proteinei este reprezentat ca o curba polinomiala ce trece prin carbonii α ai fiecarui aminoacid)
22 - Create surface (electrostatic, hidrofobicitate,...) - din "Context menu" - Create labels (la proteina sau la ligand) - din "Context menu"
23 Structural Protein Alignment: se face prin identificarea rezidurilor identice din doua proteine si calcularea translatiilor si rotatiilor ce minimizeaza RMSD (root mean standard deviation) dintre carbonii alfa ai reziduurilor identice. - se selecteaza Tools, apoi Structural Protein Alignment. Proteina "target" este proteina ce va fi translatata si reorientata (in Visualization window).
24 Energy Map Visualization: se vizualizeaza campul de forte Steric favorable: regiunile favorabile plasarii unui atom nepolar. Potential electrostatic: rosu (sarcina electrostatica negativa), albastru (sarcina electrostatica pozitiva)
25 Model activitate practica: Vizualizarea unei proteinei (si a liganzilor) folosind "Molegro Molecular Viewer" 1) proteina :
26 2) Liganzii proteinei distanta dintre atomii C(8) si O(13) din: 1,20 A: unghiul dintre 2 legaturi adiacente: (C5-Se6-C7=102,14)
27 3) vizualizarea aminoacizilor aflati la 7 Å de centrul proteinei;
28 3) aminoacizii aflati la 10 Å de centrul ligandului;
29 4) aminoacidul din pozitia 10: Glycina, Structura secundara: Coil
30 4) aminoacidul din pozitia 166: Lysina, Structura secundara: Beta sheet
31 5) structura secundara a proteinei: fara ligand; "color by residue" cu ligand; "color by structure"
32 6) Etichetarea aminoaciziilor
33 7) Crearea suprafetei proteinei din punct de vedere electrostatic din punct de vedere al hidrofobicitatii
34 8) vizualizarea (concomitenta) suprafetei proteinei (solid), a unei portiuni a lantului de aminoacizi si a ligandului:
35 9) harti energetice (1EMA) sarcina electrostatica zone favorabile steric
36 zone favorabile acceptarii atomilor de hidrogen zone favorabile donarii atomilor de hidrogen
37 Suprapunerea celor 4 reprezentari energetice Molecula in reprezentare Backbone (cartoon, color by residue)
Pymol Practial Guide
 Pymol Practial Guide Pymol is a powerful visualizor very convenient to work with protein molecules. Its interface may seem complex at first, but you will see that with a little practice is simple and powerful
Pymol Practial Guide Pymol is a powerful visualizor very convenient to work with protein molecules. Its interface may seem complex at first, but you will see that with a little practice is simple and powerful
Soluţii juniori., unde 1, 2
 Soluţii juniori Problema 1 Se consideră suma S x1x x3x4... x015 x016 Este posibil să avem S 016? Răspuns: Da., unde 1,,..., 016 3, 3 Termenii sumei sunt de forma 3 3 1, x x x. 3 5 6 sau Cristian Lazăr
Soluţii juniori Problema 1 Se consideră suma S x1x x3x4... x015 x016 Este posibil să avem S 016? Răspuns: Da., unde 1,,..., 016 3, 3 Termenii sumei sunt de forma 3 3 1, x x x. 3 5 6 sau Cristian Lazăr
Tutorial. Getting started. Sample to Insight. March 31, 2016
 Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
Getting started March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com Getting started
Subiecte geometrie licenta matematica-informatica 4 ani
 Class: Date: Subiecte geometrie licenta matematica-informatica 4 ani Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. Complementara unui subspatiu
Class: Date: Subiecte geometrie licenta matematica-informatica 4 ani Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. Complementara unui subspatiu
Molecular modeling with InsightII
 Molecular modeling with InsightII Yuk Sham Computational Biology/Biochemistry Consultant Phone: (612) 624 7427 (Walter Library) Phone: (612) 624 0783 (VWL) Email: shamy@msi.umn.edu How to run InsightII
Molecular modeling with InsightII Yuk Sham Computational Biology/Biochemistry Consultant Phone: (612) 624 7427 (Walter Library) Phone: (612) 624 0783 (VWL) Email: shamy@msi.umn.edu How to run InsightII
Protein Bioinformatics Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski
 Protein Bioinformatics 260.655 Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski Goals: Approx. Time [1] Use the Protein Data Bank PDB website. 10 minutes [2] Use the WebMol Viewer.
Protein Bioinformatics 260.655 Computer lab #1 Friday, April 11, 2008 Sean Prigge and Ingo Ruczinski Goals: Approx. Time [1] Use the Protein Data Bank PDB website. 10 minutes [2] Use the WebMol Viewer.
Part 7 Bonds and Structural Supports
 Part 7 Bonds and Structural Supports http://cbm.msoe.edu/newwebsite/learntomodel Introduction In addition to covalent bonds between atoms in a molecule, Jmol has the ability to render Hydrogen Bonds and
Part 7 Bonds and Structural Supports http://cbm.msoe.edu/newwebsite/learntomodel Introduction In addition to covalent bonds between atoms in a molecule, Jmol has the ability to render Hydrogen Bonds and
Bonds and Structural Supports
 Bonds and Structural Supports Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/struts.html
Bonds and Structural Supports Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/struts.html
Sisteme cu logica fuzzy
 Sisteme cu logica fuzzy 1/15 Sisteme cu logica fuzzy Mamdani Fie un sistem cu logică fuzzy Mamdani două intrări x şi y ieşire z x y SLF Structura z 2/15 Sisteme cu logica fuzzy Mamdani Baza de reguli R
Sisteme cu logica fuzzy 1/15 Sisteme cu logica fuzzy Mamdani Fie un sistem cu logică fuzzy Mamdani două intrări x şi y ieşire z x y SLF Structura z 2/15 Sisteme cu logica fuzzy Mamdani Baza de reguli R
SeeSAR 7.1 Beginners Guide. June 2017
 SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
SeeSAR 7.1 Beginners Guide June 2017 Part 1: Basics 1 Type a pdb code and press return or Load your own protein or already existing project, or Just load molecules To begin, let s type 2zff and download
3D - Structure Graphics Capabilities with PDF-4 Database Products
 3D - Structure Graphics Capabilities with PDF-4 Database Products Atomic Structure Information in the PDF-4 Databases ICDD s PDF-4 databases contain atomic structure information for a significant number
3D - Structure Graphics Capabilities with PDF-4 Database Products Atomic Structure Information in the PDF-4 Databases ICDD s PDF-4 databases contain atomic structure information for a significant number
Building small molecules
 Building small molecules Use the Builder (right panel) to build up molecules. Start building clicking a fragment/atom in the builder and it will appear to the workspace. Continue modifying the molecule
Building small molecules Use the Builder (right panel) to build up molecules. Start building clicking a fragment/atom in the builder and it will appear to the workspace. Continue modifying the molecule
Silica surface - Materials Studio tutorial. CREATING SiO 2 SURFACE
 Silica surface - Materials Studio tutorial CREATING SiO 2 SURFACE Our goal surface of SiO2 6.948 Ǻ Import structure The XRD experiment gives us such parameters as: lattice parameters, symmetry group and
Silica surface - Materials Studio tutorial CREATING SiO 2 SURFACE Our goal surface of SiO2 6.948 Ǻ Import structure The XRD experiment gives us such parameters as: lattice parameters, symmetry group and
Examples of Protein Modeling. Protein Modeling. Primary Structure. Protein Structure Description. Protein Sequence Sources. Importing Sequences to MOE
 Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
2.1-PORNIREA, REPORNIREA SI OPRIREA CALCULATORULUI 2.2.-VIZUALIZAREA INFORMAŢIILOR DESPRE RESURSELE HARDWARE SI SOFTWARE
 2.1-PORNIREA, REPORNIREA SI OPRIREA CALCULATORULUI 2.2.-VIZUALIZAREA INFORMAŢIILOR DESPRE RESURSELE HARDWARE SI SOFTWARE 2.3.-DESCRIEREA ŞI PERSONALIZAREA INTERFEŢEI SISTEMULUI DE OPERARE DESCRIEREA SI
2.1-PORNIREA, REPORNIREA SI OPRIREA CALCULATORULUI 2.2.-VIZUALIZAREA INFORMAŢIILOR DESPRE RESURSELE HARDWARE SI SOFTWARE 2.3.-DESCRIEREA ŞI PERSONALIZAREA INTERFEŢEI SISTEMULUI DE OPERARE DESCRIEREA SI
Pentru clasa a X-a Ştiinţele naturii-sem II
 Pentru clasa a X-a Ştiinţele naturii-sem II Reprezentarea algoritmilor. Pseudocod. Principiile programării structurate. Structuri de bază: structura liniară structura alternativă structura repetitivă Algoritmi
Pentru clasa a X-a Ştiinţele naturii-sem II Reprezentarea algoritmilor. Pseudocod. Principiile programării structurate. Structuri de bază: structura liniară structura alternativă structura repetitivă Algoritmi
Section III - Designing Models for 3D Printing
 Section III - Designing Models for 3D Printing In this section of the Jmol Training Guide, you will become familiar with the commands needed to design a model that will be built on a 3D Printer. As you
Section III - Designing Models for 3D Printing In this section of the Jmol Training Guide, you will become familiar with the commands needed to design a model that will be built on a 3D Printer. As you
Part 8 Working with Nucleic Acids
 Part 8 Working with Nucleic Acids http://cbm.msoe.edu/newwebsite/learntomodel Introduction Most Protein Databank files loaded into the CBM's Jmol Design Environment include protein structures and small
Part 8 Working with Nucleic Acids http://cbm.msoe.edu/newwebsite/learntomodel Introduction Most Protein Databank files loaded into the CBM's Jmol Design Environment include protein structures and small
Inteligenta Artificiala
 Inteligenta Artificiala Universitatea Politehnica Bucuresti Anul universitar 2010-2011 Adina Magda Florea http://turing.cs.pub.ro/ia_10 si curs.cs.pub.ro 1 Curs nr. 4 Cautare cu actiuni nedeterministe
Inteligenta Artificiala Universitatea Politehnica Bucuresti Anul universitar 2010-2011 Adina Magda Florea http://turing.cs.pub.ro/ia_10 si curs.cs.pub.ro 1 Curs nr. 4 Cautare cu actiuni nedeterministe
Tutorial: Structural Analysis of a Protein-Protein Complex
 Molecular Modeling Section (MMS) Department of Pharmaceutical and Pharmacological Sciences University of Padova Via Marzolo 5-35131 Padova (IT) @contact: stefano.moro@unipd.it Tutorial: Structural Analysis
Molecular Modeling Section (MMS) Department of Pharmaceutical and Pharmacological Sciences University of Padova Via Marzolo 5-35131 Padova (IT) @contact: stefano.moro@unipd.it Tutorial: Structural Analysis
Introduction to Structure Preparation and Visualization
 Introduction to Structure Preparation and Visualization Created with: Release 2018-4 Prerequisites: Release 2018-2 or higher Access to the internet Categories: Molecular Visualization, Structure-Based
Introduction to Structure Preparation and Visualization Created with: Release 2018-4 Prerequisites: Release 2018-2 or higher Access to the internet Categories: Molecular Visualization, Structure-Based
The Structure and Functions of Proteins
 Wright State University CORE Scholar Computer Science and Engineering Faculty Publications Computer Science and Engineering 2003 The Structure and Functions of Proteins Dan E. Krane Wright State University
Wright State University CORE Scholar Computer Science and Engineering Faculty Publications Computer Science and Engineering 2003 The Structure and Functions of Proteins Dan E. Krane Wright State University
BUILDING BASICS WITH HYPERCHEM LITE
 BUILDING BASICS WITH HYPERCHEM LITE LAB MOD1.COMP From Gannon University SIM INTRODUCTION A chemical bond is a link between atoms resulting from the mutual attraction of their nuclei for electrons. There
BUILDING BASICS WITH HYPERCHEM LITE LAB MOD1.COMP From Gannon University SIM INTRODUCTION A chemical bond is a link between atoms resulting from the mutual attraction of their nuclei for electrons. There
SOI prin smart-cut. Caracterizarea TEM-HRTEM a defectelor structuale induse in Si prin hidrogenare in plasma.
 SOI prin smart-cut. Caracterizarea TEM-HRTEM a defectelor structuale induse in Si prin hidrogenare in plasma. Dr. Corneliu GHICA, Dr. Leona NISTOR Proiect IDEI, Contract Nr. 233/2007 1. C. Ghica, L. C.
SOI prin smart-cut. Caracterizarea TEM-HRTEM a defectelor structuale induse in Si prin hidrogenare in plasma. Dr. Corneliu GHICA, Dr. Leona NISTOR Proiect IDEI, Contract Nr. 233/2007 1. C. Ghica, L. C.
Programarea Dinamica. (si alte chestii adiacente) Andrei Olariu
 Programarea Dinamica (si alte chestii adiacente) Andrei Olariu andrei@olariu.org Despre mine - Absolvent FMI UniBuc - Doctorand in prelucrarea limbajului natural, in special in mediul online (Twitter)
Programarea Dinamica (si alte chestii adiacente) Andrei Olariu andrei@olariu.org Despre mine - Absolvent FMI UniBuc - Doctorand in prelucrarea limbajului natural, in special in mediul online (Twitter)
O V E R V I E W. This study suggests grouping of numbers that do not divide the number
 MSCN(2010) : 11A99 Author : Barar Stelian Liviu Adress : Israel e-mail : stelibarar@yahoo.com O V E R V I E W This study suggests grouping of numbers that do not divide the number 3 and/or 5 in eight collumns.
MSCN(2010) : 11A99 Author : Barar Stelian Liviu Adress : Israel e-mail : stelibarar@yahoo.com O V E R V I E W This study suggests grouping of numbers that do not divide the number 3 and/or 5 in eight collumns.
4/68. Mini-comutatoare cu came. Prezentare generalã a sistemului. Întreruptoare Pornit-Oprit TM. Comutatoare de comandã TM.
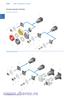 / Mini-comutatoare cu came Prezentare generalã a sistemului Întreruptoare Pornit-Oprit Comutatoare de comandã HA ND AU TO HPL-de-DE O Mini-comutatoare cu came / Montaj pe ușã (.../E) Frontal IP Montaj
/ Mini-comutatoare cu came Prezentare generalã a sistemului Întreruptoare Pornit-Oprit Comutatoare de comandã HA ND AU TO HPL-de-DE O Mini-comutatoare cu came / Montaj pe ușã (.../E) Frontal IP Montaj
STRUCTURAL INTENSITY METHOD APPLIED TO STUDY OF VIBRATIONS DAMPING / METODA INTENSIMETRIEI STUCTURALE APLICATĂ LA STUDIUL AMORTIZĂRII VIBRAŢIILOR
 Vol.48, No. / 06 STRUCTURAL INTENSITY METHOD APPLIED TO STUDY OF VIBRATIONS DAMPING / METODA INTENSIMETRIEI STUCTURALE APLICATĂ LA STUDIUL AMORTIZĂRII VIBRAŢIILOR Assoc. Prof. Ph.D. Eng. Carp-Ciocârdia
Vol.48, No. / 06 STRUCTURAL INTENSITY METHOD APPLIED TO STUDY OF VIBRATIONS DAMPING / METODA INTENSIMETRIEI STUCTURALE APLICATĂ LA STUDIUL AMORTIZĂRII VIBRAŢIILOR Assoc. Prof. Ph.D. Eng. Carp-Ciocârdia
Reactoare chimice cu curgere piston (ideala) cu amestecare completa de tip batch (autoclava)
 Reactoare chimice cu curgere piston (ideala) cu amestecare completa de tip batch (autoclava) Reactorul cu curgere ideala Toate particulele se deplaseaza intr-o directie de-a lungul reactorului, precum
Reactoare chimice cu curgere piston (ideala) cu amestecare completa de tip batch (autoclava) Reactorul cu curgere ideala Toate particulele se deplaseaza intr-o directie de-a lungul reactorului, precum
Performing a Pharmacophore Search using CSD-CrossMiner
 Table of Contents Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Searching with a Pharmacophore... 4 Performing a Pharmacophore Search using CSD-CrossMiner Version 2.0
Table of Contents Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Searching with a Pharmacophore... 4 Performing a Pharmacophore Search using CSD-CrossMiner Version 2.0
Liste. Stive. Cozi SD 2017/2018
 Liste. Stive. Cozi SD 2017/2018 Conţinut Tipurile abstracte LLin, LLinOrd, Stiva, Coada Liste liniare Implementarea cu tablouri Implementarea cu liste simplu înlănțuite Liste liniare ordonate Stive Cozi
Liste. Stive. Cozi SD 2017/2018 Conţinut Tipurile abstracte LLin, LLinOrd, Stiva, Coada Liste liniare Implementarea cu tablouri Implementarea cu liste simplu înlănțuite Liste liniare ordonate Stive Cozi
Introduction Molecular Structure Script Console External resources Advanced topics. JMol tutorial. Giovanni Morelli.
 Gen 19th, 2017 1 2 Create and edit Display and view Mesurament and labelling Surface and Orbitals 3 4 from Database Protein Enzyme Crystal Structure and Unit Cell 5 Symmetry Animation General information
Gen 19th, 2017 1 2 Create and edit Display and view Mesurament and labelling Surface and Orbitals 3 4 from Database Protein Enzyme Crystal Structure and Unit Cell 5 Symmetry Animation General information
Dock Ligands from a 2D Molecule Sketch
 Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Dock Ligands from a 2D Molecule Sketch March 31, 2016 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com support-clcbio@qiagen.com
Teorema Reziduurilor şi Bucuria Integralelor Reale Prezentare de Alexandru Negrescu
 Teorema Reiduurilor şi Bucuria Integralelor Reale Preentare de Alexandru Negrescu Integrale cu funcţii raţionale ce depind de sint şi cost u notaţia e it, avem: cost sint i ( + ( dt d i, iar integrarea
Teorema Reiduurilor şi Bucuria Integralelor Reale Preentare de Alexandru Negrescu Integrale cu funcţii raţionale ce depind de sint şi cost u notaţia e it, avem: cost sint i ( + ( dt d i, iar integrarea
Viewing and Analyzing Proteins, Ligands and their Complexes 2
 2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
Dihedral Angles. Homayoun Valafar. Department of Computer Science and Engineering, USC 02/03/10 CSCE 769
 Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
Let s continue our discussion on the interaction between Fe(III) and 6,7-dihydroxynaphthalene-2- sulfonate.
 Chemistry 5995(133)-8990(013) Bioinorganic Chemistry: The Good, the Bad, and the Potential of Metals Assignment 2- Aqueous Speciation, Magnetism, Redox, UV-Vis Spectroscopy, and Pymol Let s continue our
Chemistry 5995(133)-8990(013) Bioinorganic Chemistry: The Good, the Bad, and the Potential of Metals Assignment 2- Aqueous Speciation, Magnetism, Redox, UV-Vis Spectroscopy, and Pymol Let s continue our
Working with protein structures. Benjamin Jack
 Working with protein structures Benjamin Jack Structure of Triosephosphate Isomerase PDB ID: 1HTI loop beta sheet alpha helix Different perspectives of the same structure Structure of Truncated Hemoglobin
Working with protein structures Benjamin Jack Structure of Triosephosphate Isomerase PDB ID: 1HTI loop beta sheet alpha helix Different perspectives of the same structure Structure of Truncated Hemoglobin
Contents. Acknowledgements. Section 1: Using Jmol as a Computer Visualization Tool
 1ZAA.pdb 1ZAA.pdb Contents Acknowledgements Section 1: Using Jmol as a Computer Visualization Tool 1-1 - Downloading Jmol 1-2 - Protein Data Bank (PDB) Files 1-3 - Launching Jmol and Opening a PDB File
1ZAA.pdb 1ZAA.pdb Contents Acknowledgements Section 1: Using Jmol as a Computer Visualization Tool 1-1 - Downloading Jmol 1-2 - Protein Data Bank (PDB) Files 1-3 - Launching Jmol and Opening a PDB File
Molecular Visualization. Introduction
 Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Protein Structure Basics
 Protein Structure Basics Presented by Alison Fraser, Christine Lee, Pradhuman Jhala, Corban Rivera Importance of Proteins Muscle structure depends on protein-protein interactions Transport across membranes
Protein Structure Basics Presented by Alison Fraser, Christine Lee, Pradhuman Jhala, Corban Rivera Importance of Proteins Muscle structure depends on protein-protein interactions Transport across membranes
Divizibilitate în mulțimea numerelor naturale/întregi
 Divizibilitate în mulțimea numerelor naturale/întregi Teorema îmărţirii cu rest în mulțimea numerelor naturale Fie a, b, b 0. Atunci există q, r astfel încât a=bq+r, cu 0 r < b. În lus, q şi r sunt unic
Divizibilitate în mulțimea numerelor naturale/întregi Teorema îmărţirii cu rest în mulțimea numerelor naturale Fie a, b, b 0. Atunci există q, r astfel încât a=bq+r, cu 0 r < b. În lus, q şi r sunt unic
From Amino Acids to Proteins - in 4 Easy Steps
 From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
Basics of protein structure
 Today: 1. Projects a. Requirements: i. Critical review of one paper ii. At least one computational result b. Noon, Dec. 3 rd written report and oral presentation are due; submit via email to bphys101@fas.harvard.edu
Today: 1. Projects a. Requirements: i. Critical review of one paper ii. At least one computational result b. Noon, Dec. 3 rd written report and oral presentation are due; submit via email to bphys101@fas.harvard.edu
PRELUCRARI PE IMAGINI BINARE (ALB/NEGRU)
 PRELUCRRI PE IMGINI BINRE (LB/NEGRU) Imagine binara? 2 nuante: alb ( 0 ) pixelii de fond ( I(x,y)= 255 pt. imagini indexate cu 8 biti/pixel ) negru ( 1 ) pixelii apartinand obiectelor ( I(x,y)= 0 pt. imagini
PRELUCRRI PE IMGINI BINRE (LB/NEGRU) Imagine binara? 2 nuante: alb ( 0 ) pixelii de fond ( I(x,y)= 255 pt. imagini indexate cu 8 biti/pixel ) negru ( 1 ) pixelii apartinand obiectelor ( I(x,y)= 0 pt. imagini
CAP 5510 Lecture 3 Protein Structures
 CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
Rezolvarea ecuaţiilor şi sistemelor de ecuaţii diferenţiale ordinare (II)
 Rezolvarea ecuaţiilor şi sistemelor de ecuaţii diferenţiale ordinare (II) Metode multipas Prof.dr.ing. Universitatea "Politehnica" Bucureşti, Facultatea de Inginerie Electrică Suport didactic pentru disciplina
Rezolvarea ecuaţiilor şi sistemelor de ecuaţii diferenţiale ordinare (II) Metode multipas Prof.dr.ing. Universitatea "Politehnica" Bucureşti, Facultatea de Inginerie Electrică Suport didactic pentru disciplina
CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis
 CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis Purpose: The purpose of this laboratory is to introduce some of the basic visualization and modeling tools for viewing
CHEM 463: Advanced Inorganic Chemistry Modeling Metalloproteins for Structural Analysis Purpose: The purpose of this laboratory is to introduce some of the basic visualization and modeling tools for viewing
Matematici speciale Integrarea functiilor complexe
 Matematii speiale Integrarea funtiilor omplexe Martie 18 ii Be yourself, everyone else is already taken. Osar Wilde 5 Integrarea funtiilor omplexe Integrala Riemann a unei funtii u valori omplexe se defineste
Matematii speiale Integrarea funtiilor omplexe Martie 18 ii Be yourself, everyone else is already taken. Osar Wilde 5 Integrarea funtiilor omplexe Integrala Riemann a unei funtii u valori omplexe se defineste
Protein structure (and biomolecular structure more generally) CS/CME/BioE/Biophys/BMI 279 Sept. 28 and Oct. 3, 2017 Ron Dror
 Protein structure (and biomolecular structure more generally) CS/CME/BioE/Biophys/BMI 279 Sept. 28 and Oct. 3, 2017 Ron Dror Please interrupt if you have questions, and especially if you re confused! Assignment
Protein structure (and biomolecular structure more generally) CS/CME/BioE/Biophys/BMI 279 Sept. 28 and Oct. 3, 2017 Ron Dror Please interrupt if you have questions, and especially if you re confused! Assignment
Orientational degeneracy in the presence of one alignment tensor.
 Orientational degeneracy in the presence of one alignment tensor. Rotation about the x, y and z axes can be performed in the aligned mode of the program to examine the four degenerate orientations of two
Orientational degeneracy in the presence of one alignment tensor. Rotation about the x, y and z axes can be performed in the aligned mode of the program to examine the four degenerate orientations of two
12 Molecular Visualization of an Enzyme, Acetylcholinesterase
 12 Molecular Visualization of an Enzyme, Acetylcholinesterase 12.1 Learning Objectives The goal of this laboratory is to analyze some of the major structural elements of an important enzyme, acetylcholinesterase
12 Molecular Visualization of an Enzyme, Acetylcholinesterase 12.1 Learning Objectives The goal of this laboratory is to analyze some of the major structural elements of an important enzyme, acetylcholinesterase
Preparing a PDB File
 Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
Figure 1: Schematic view of the ligand-binding domain from the vitamin D receptor (PDB file 1IE9). The crystallographic waters are shown as small spheres and the bound ligand is shown as a CPK model. HO
COMPARATIVE DISCUSSION ABOUT THE DETERMINING METHODS OF THE STRESSES IN PLANE SLABS
 74 COMPARATIVE DISCUSSION ABOUT THE DETERMINING METHODS OF THE STRESSES IN PLANE SLABS Codrin PRECUPANU 3, Dan PRECUPANU,, Ștefan OPREA Correspondent Member of Technical Sciences Academy Gh. Asachi Technical
74 COMPARATIVE DISCUSSION ABOUT THE DETERMINING METHODS OF THE STRESSES IN PLANE SLABS Codrin PRECUPANU 3, Dan PRECUPANU,, Ștefan OPREA Correspondent Member of Technical Sciences Academy Gh. Asachi Technical
BOND LENGTH WITH HYPERCHEM LITE
 BOND LENGTH WITH HYPERCHEM LITE LAB MOD2.COMP From Gannon University SIM INTRODUCTION The electron cloud surrounding the nucleus of the atom determines the size of the atom. Since this distance is somewhat
BOND LENGTH WITH HYPERCHEM LITE LAB MOD2.COMP From Gannon University SIM INTRODUCTION The electron cloud surrounding the nucleus of the atom determines the size of the atom. Since this distance is somewhat
Creating a Pharmacophore Query from a Reference Molecule & Scaffold Hopping in CSD-CrossMiner
 Table of Contents Creating a Pharmacophore Query from a Reference Molecule & Scaffold Hopping in CSD-CrossMiner Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Features
Table of Contents Creating a Pharmacophore Query from a Reference Molecule & Scaffold Hopping in CSD-CrossMiner Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Features
Figure 1. Indinavir. Most of the hydrogens have been left off to make the structure easier to see. We will focus on just the central OH group.
 1 Experiment 11: Molecular Mechanics and Modeling Purpose: ne purpose is to have some fun learning about molecular modeling and mechanics. The other purpose is to begin to appreciate how complex molecules
1 Experiment 11: Molecular Mechanics and Modeling Purpose: ne purpose is to have some fun learning about molecular modeling and mechanics. The other purpose is to begin to appreciate how complex molecules
Utilizarea limbajului SQL pentru cereri OLAP. Mihaela Muntean 2015
 Utilizarea limbajului SQL pentru cereri OLAP Mihaela Muntean 2015 Cuprins Implementarea operatiilor OLAP de baza in SQL -traditional: Rollup Slice Dice Pivotare SQL-2008 Optiunea ROLLUP Optiunea CUBE,
Utilizarea limbajului SQL pentru cereri OLAP Mihaela Muntean 2015 Cuprins Implementarea operatiilor OLAP de baza in SQL -traditional: Rollup Slice Dice Pivotare SQL-2008 Optiunea ROLLUP Optiunea CUBE,
Get familiar with PDBsum and the PDB Extract atomic coordinates from protein data files Compute bond angles and dihedral angles
 CS483 Assignment #2 Due date: Mar. 1 at the start of class. Protein Geometry Bedbug spit? Just say NO! Purpose of this assignment Get familiar with PDBsum and the PDB Extract atomic coordinates from protein
CS483 Assignment #2 Due date: Mar. 1 at the start of class. Protein Geometry Bedbug spit? Just say NO! Purpose of this assignment Get familiar with PDBsum and the PDB Extract atomic coordinates from protein
Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase Cwc27
 Acta Cryst. (2014). D70, doi:10.1107/s1399004714021695 Supporting information Volume 70 (2014) Supporting information for article: Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase
Acta Cryst. (2014). D70, doi:10.1107/s1399004714021695 Supporting information Volume 70 (2014) Supporting information for article: Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase
Visualization of Macromolecular Structures
 Visualization of Macromolecular Structures Present by: Qihang Li orig. author: O Donoghue, et al. Structural biology is rapidly accumulating a wealth of detailed information. Over 60,000 high-resolution
Visualization of Macromolecular Structures Present by: Qihang Li orig. author: O Donoghue, et al. Structural biology is rapidly accumulating a wealth of detailed information. Over 60,000 high-resolution
Molecular Modeling and Conformational Analysis with PC Spartan
 Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Molecular Modeling and Conformational Analysis with PC Spartan Introduction Molecular modeling can be done in a variety of ways, from using simple hand-held models to doing sophisticated calculations on
Introducing Hippy: A visualization tool for understanding the α-helix pair interface
 Introducing Hippy: A visualization tool for understanding the α-helix pair interface Robert Fraser and Janice Glasgow School of Computing, Queen s University, Kingston ON, Canada, K7L3N6 {robert,janice}@cs.queensu.ca
Introducing Hippy: A visualization tool for understanding the α-helix pair interface Robert Fraser and Janice Glasgow School of Computing, Queen s University, Kingston ON, Canada, K7L3N6 {robert,janice}@cs.queensu.ca
SUPPLEMENTARY INFORMATION
 Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
Supplementary Results DNA binding property of the SRA domain was examined by an electrophoresis mobility shift assay (EMSA) using synthesized 12-bp oligonucleotide duplexes containing unmodified, hemi-methylated,
The Select Command and Boolean Operators
 The Select Command and Boolean Operators Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/boolean.html
The Select Command and Boolean Operators Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/boolean.html
Part 4 The Select Command and Boolean Operators
 Part 4 The Select Command and Boolean Operators http://cbm.msoe.edu/newwebsite/learntomodel Introduction By default, every command you enter into the Console affects the entire molecular structure. However,
Part 4 The Select Command and Boolean Operators http://cbm.msoe.edu/newwebsite/learntomodel Introduction By default, every command you enter into the Console affects the entire molecular structure. However,
Right click on the link and save the file on the disk (Save link target as...). Then execute this in the command window:
 Läkemedelsutveckling ht 2005 Copyright 2005 Lars Brive Excercise Analysis of structures of protein ligand complexes In this excercise you will examine the geometrical features of six x ray structures in
Läkemedelsutveckling ht 2005 Copyright 2005 Lars Brive Excercise Analysis of structures of protein ligand complexes In this excercise you will examine the geometrical features of six x ray structures in
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis Sergio de Cima, Luis M. Polo, Carmen Díez-Fernández, Ana I. Martínez, Javier
SUPPLEMENTARY INFORMATION Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis Sergio de Cima, Luis M. Polo, Carmen Díez-Fernández, Ana I. Martínez, Javier
COMPARATIVE STUDY OF STRUCTURAL ANALYSIS APPLIED TO AGRICULTURAL MACHINES BODIES AND ACCOMPLISHED WITH SOLID WORKS AND AUTODESK INVENTOR PROGRAMS
 COMPARATIVE STUDY OF STRUCTURAL ANALYSIS APPLIED TO AGRICULTURAL MACHINES BODIES AND ACCOMPLISHED WITH SOLID WORKS AND AUTODESK INVENTOR PROGRAMS / STUDIU COMPARATIV DE ANALIZA STRUCTURALA APLICATA LA
COMPARATIVE STUDY OF STRUCTURAL ANALYSIS APPLIED TO AGRICULTURAL MACHINES BODIES AND ACCOMPLISHED WITH SOLID WORKS AND AUTODESK INVENTOR PROGRAMS / STUDIU COMPARATIV DE ANALIZA STRUCTURALA APLICATA LA
Gradul de comutativitate al grupurilor finite 1
 Gradul de comutativitate al grupurilor finite Marius TĂRNĂUCEANU Abstract The commutativity degree of a group is one of the most important probabilistic aspects of finite group theory In this survey we
Gradul de comutativitate al grupurilor finite Marius TĂRNĂUCEANU Abstract The commutativity degree of a group is one of the most important probabilistic aspects of finite group theory In this survey we
FINDING THE TRACES OF A GIVEN PLANE: ANALYTICALLY AND THROUGH GRAPHICAL CONSTRUCTIONS
 BULETINUL INSTITUTULUI POLITEHNI DIN IŞI Publicat de Universitatea Tehnică Gheorghe sachi din Iaşi Tomul LVII (LXI), Fasc. 3, 20 Secţia ONSTRUŢII DE MŞINI FINDING THE TRES OF GIVEN PLNE: NLYTILLY ND THROUGH
BULETINUL INSTITUTULUI POLITEHNI DIN IŞI Publicat de Universitatea Tehnică Gheorghe sachi din Iaşi Tomul LVII (LXI), Fasc. 3, 20 Secţia ONSTRUŢII DE MŞINI FINDING THE TRES OF GIVEN PLNE: NLYTILLY ND THROUGH
BCMP 201 Protein biochemistry
 BCMP 201 Protein biochemistry BCMP 201 Protein biochemistry with emphasis on the interrelated roles of protein structure, catalytic activity, and macromolecular interactions in biological processes. The
BCMP 201 Protein biochemistry BCMP 201 Protein biochemistry with emphasis on the interrelated roles of protein structure, catalytic activity, and macromolecular interactions in biological processes. The
3D Molecule Viewer of MOGADOC (JavaScript)
 3D Molecule Viewer of MOGADOC (JavaScript) Movement of the Molecule Rotation of the molecule: Use left mouse button to drag. Translation of the molecule: Use right mouse button to drag. Resize the molecule:
3D Molecule Viewer of MOGADOC (JavaScript) Movement of the Molecule Rotation of the molecule: Use left mouse button to drag. Translation of the molecule: Use right mouse button to drag. Resize the molecule:
MINERALOGIE NOTE DE CURS
 MINERALOGIE NOTE DE CURS 1 CUPRINS I MINERALOGIE GENERALA 5 1. INTRODUCERE 5 2. PRINCIPII DE FORMARE A STRUCTURII MINERALE 6 2.1. Stabilitatea structurii minerale 2.2. Legaturi chimice. Dimensiunile efective
MINERALOGIE NOTE DE CURS 1 CUPRINS I MINERALOGIE GENERALA 5 1. INTRODUCERE 5 2. PRINCIPII DE FORMARE A STRUCTURII MINERALE 6 2.1. Stabilitatea structurii minerale 2.2. Legaturi chimice. Dimensiunile efective
Exercises for Windows
 Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Exercises for Windows CAChe User Interface for Windows Select tool Application window Document window (workspace) Style bar Tool palette Select entire molecule Select Similar Group Select Atom tool Rotate
Protein Structures. Sequences of amino acid residues 20 different amino acids. Quaternary. Primary. Tertiary. Secondary. 10/8/2002 Lecture 12 1
 Protein Structures Sequences of amino acid residues 20 different amino acids Primary Secondary Tertiary Quaternary 10/8/2002 Lecture 12 1 Angles φ and ψ in the polypeptide chain 10/8/2002 Lecture 12 2
Protein Structures Sequences of amino acid residues 20 different amino acids Primary Secondary Tertiary Quaternary 10/8/2002 Lecture 12 1 Angles φ and ψ in the polypeptide chain 10/8/2002 Lecture 12 2
Design of a Novel Globular Protein Fold with Atomic-Level Accuracy
 Design of a Novel Globular Protein Fold with Atomic-Level Accuracy Brian Kuhlman, Gautam Dantas, Gregory C. Ireton, Gabriele Varani, Barry L. Stoddard, David Baker Presented by Kate Stafford 4 May 05 Protein
Design of a Novel Globular Protein Fold with Atomic-Level Accuracy Brian Kuhlman, Gautam Dantas, Gregory C. Ireton, Gabriele Varani, Barry L. Stoddard, David Baker Presented by Kate Stafford 4 May 05 Protein
Barem de notare clasa a V-a
 Barem de notare clasa a V-a Problema1. Determinați mulțimile A și B, formate din numere naturale, știind că îndeplinesc simultan condițiile: a) A B,5,6 ; b) B A 0,7 ; c) card AB 3; d) suma elementelor
Barem de notare clasa a V-a Problema1. Determinați mulțimile A și B, formate din numere naturale, știind că îndeplinesc simultan condițiile: a) A B,5,6 ; b) B A 0,7 ; c) card AB 3; d) suma elementelor
ON THE QUATERNARY QUADRATIC DIOPHANTINE EQUATIONS (II) NICOLAE BRATU 1 ADINA CRETAN 2
 ON THE QUATERNARY QUADRATIC DIOPHANTINE EQUATIONS (II) NICOLAE BRATU 1 ADINA CRETAN ABSTRACT This paper has been updated and completed thanks to suggestions and critics coming from Dr. Mike Hirschhorn,
ON THE QUATERNARY QUADRATIC DIOPHANTINE EQUATIONS (II) NICOLAE BRATU 1 ADINA CRETAN ABSTRACT This paper has been updated and completed thanks to suggestions and critics coming from Dr. Mike Hirschhorn,
Project Manual Bio3055. Apoptosis: Caspase-1
 Project Manual Bio3055 Apoptosis: Caspase-1 Bednarski 2003 Funded by HHMI Apoptosis: Caspase-1 Introduction: Apoptosis is another name for programmed cell death. It is a series of events in a cell that
Project Manual Bio3055 Apoptosis: Caspase-1 Bednarski 2003 Funded by HHMI Apoptosis: Caspase-1 Introduction: Apoptosis is another name for programmed cell death. It is a series of events in a cell that
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
BIBC 100. Structural Biochemistry
 BIBC 100 Structural Biochemistry http://classes.biology.ucsd.edu/bibc100.wi14 Papers- Dialogue with Scientists Questions: Why? How? What? So What? Dialogue Structure to explain function Knowledge Food
BIBC 100 Structural Biochemistry http://classes.biology.ucsd.edu/bibc100.wi14 Papers- Dialogue with Scientists Questions: Why? How? What? So What? Dialogue Structure to explain function Knowledge Food
"IIITO-TEC 'NIKI" & EQUIPME
 LIGHTING "IIITO-TEC 'NIKI" & EQUIPME T FOR CITIES 6 MAKEDONOMAHON STR.,ZIPCaDE:67009,KALO ORI,THESSALONIKI, GREECE TEL / FAX: 0030 2310761824/751626,8 mall: hito@otenet.qi' Webslte:www.hlto..techkl.gr
LIGHTING "IIITO-TEC 'NIKI" & EQUIPME T FOR CITIES 6 MAKEDONOMAHON STR.,ZIPCaDE:67009,KALO ORI,THESSALONIKI, GREECE TEL / FAX: 0030 2310761824/751626,8 mall: hito@otenet.qi' Webslte:www.hlto..techkl.gr
Ligand Scout Tutorials
 Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Ligand Scout Tutorials Step : Creating a pharmacophore from a protein-ligand complex. Type ke6 in the upper right area of the screen and press the button Download *+. The protein will be downloaded and
Molecular Modeling lecture 2
 Molecular Modeling 2018 -- lecture 2 Topics 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction 4. Where do protein structures come from? X-ray crystallography
Molecular Modeling 2018 -- lecture 2 Topics 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction 4. Where do protein structures come from? X-ray crystallography
ScienceWord and PagePlayer Chemical formulae. Dr Emile C. B. COMLAN Novoasoft Representative in Africa
 ScienceWord and PagePlayer Chemical formulae Dr Emile C. B. CMLAN Novoasoft Representative in Africa Emails: ecomlan@yahoo.com ecomlan@scienceoffice.com Web site: www.scienceoffice.com Chemical formulae
ScienceWord and PagePlayer Chemical formulae Dr Emile C. B. CMLAN Novoasoft Representative in Africa Emails: ecomlan@yahoo.com ecomlan@scienceoffice.com Web site: www.scienceoffice.com Chemical formulae
NMR, X-ray Diffraction, Protein Structure, and RasMol
 NMR, X-ray Diffraction, Protein Structure, and RasMol Introduction So far we have been mostly concerned with the proteins themselves. The techniques (NMR or X-ray diffraction) used to determine a structure
NMR, X-ray Diffraction, Protein Structure, and RasMol Introduction So far we have been mostly concerned with the proteins themselves. The techniques (NMR or X-ray diffraction) used to determine a structure
ATTENUATION OF THE ACOUSTIC SCREENS IN CLOSED SPACES
 U.P.B. Sci. Bull., Series D, Vol. 69, No. 3, 007 ISSN 15-358 ATTENUATION OF THE ACOUSTIC SCREENS IN CLOSED SPACES Ioan MAGHEŢI 1, Mariana SAVU Lucrarea prezintă calculul atenuării acustice a unui ecran
U.P.B. Sci. Bull., Series D, Vol. 69, No. 3, 007 ISSN 15-358 ATTENUATION OF THE ACOUSTIC SCREENS IN CLOSED SPACES Ioan MAGHEŢI 1, Mariana SAVU Lucrarea prezintă calculul atenuării acustice a unui ecran
SAM Teacher s Guide Protein Partnering and Function
 SAM Teacher s Guide Protein Partnering and Function Overview Students explore protein molecules physical and chemical characteristics and learn that these unique characteristics enable other molecules
SAM Teacher s Guide Protein Partnering and Function Overview Students explore protein molecules physical and chemical characteristics and learn that these unique characteristics enable other molecules
APBS electrostatics in VMD - Software. APBS! >!Examples! >!Visualization! >! Contents
 Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
CHEM 4170 Problem Set #1
 CHEM 4170 Problem Set #1 0. Work problems 1-7 at the end of Chapter ne and problems 1, 3, 4, 5, 8, 10, 12, 17, 18, 19, 22, 24, and 25 at the end of Chapter Two and problem 1 at the end of Chapter Three
CHEM 4170 Problem Set #1 0. Work problems 1-7 at the end of Chapter ne and problems 1, 3, 4, 5, 8, 10, 12, 17, 18, 19, 22, 24, and 25 at the end of Chapter Two and problem 1 at the end of Chapter Three
Useful background reading
 Overview of lecture * General comment on peptide bond * Discussion of backbone dihedral angles * Discussion of Ramachandran plots * Description of helix types. * Description of structures * NMR patterns
Overview of lecture * General comment on peptide bond * Discussion of backbone dihedral angles * Discussion of Ramachandran plots * Description of helix types. * Description of structures * NMR patterns
STRUCTURAL BIOINFORMATICS. Barry Grant University of Michigan
 STRUCTURAL BIOINFORMATICS Barry Grant University of Michigan www.thegrantlab.org bjgrant@umich.edu Bergen, Norway 28-Sep-2015 Objective: Provide an introduction to the practice of structural bioinformatics,
STRUCTURAL BIOINFORMATICS Barry Grant University of Michigan www.thegrantlab.org bjgrant@umich.edu Bergen, Norway 28-Sep-2015 Objective: Provide an introduction to the practice of structural bioinformatics,
Dana Alsulaibi. Jaleel G.Sweis. Mamoon Ahram
 15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
Figura 7.12 Multiscopul: schema bloc simplificată a părţii specifice osciloscopului hibrid. U Y CS S/T-H ADC MD DAC TC
 7-7 7.3.3 OSCILOSCOPUL HIBRID CE GP-IB ADC Frecvenţmetru Fazmetru Generator de caractere X Y Z Elemente de comandă şi reglaj Figura 7.1 Multiscopul: schema bloc simplificată a părţii specifice osciloscopului
7-7 7.3.3 OSCILOSCOPUL HIBRID CE GP-IB ADC Frecvenţmetru Fazmetru Generator de caractere X Y Z Elemente de comandă şi reglaj Figura 7.1 Multiscopul: schema bloc simplificată a părţii specifice osciloscopului
Protein Structures. 11/19/2002 Lecture 24 1
 Protein Structures 11/19/2002 Lecture 24 1 All 3 figures are cartoons of an amino acid residue. 11/19/2002 Lecture 24 2 Peptide bonds in chains of residues 11/19/2002 Lecture 24 3 Angles φ and ψ in the
Protein Structures 11/19/2002 Lecture 24 1 All 3 figures are cartoons of an amino acid residue. 11/19/2002 Lecture 24 2 Peptide bonds in chains of residues 11/19/2002 Lecture 24 3 Angles φ and ψ in the
Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 Å resolution: comparison with the Azotobacter vinelandii MoFe protein
 Acta Cryst. (2015). D71, 274-282, doi:10.1107/s1399004714025243 Supporting information Volume 71 (2015) Supporting information for article: Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08
Acta Cryst. (2015). D71, 274-282, doi:10.1107/s1399004714025243 Supporting information Volume 71 (2015) Supporting information for article: Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08
LIGHTNING MVP System
 LIGHTNING MVP System Lightning MVP System Control (HACCP+SSOP) Swab-uri pentru lichide si pentru Suprafete Accesorii ph Temperatura Condutivitate Monitorizare ATP Prin Bioluminescenta Cel mai complet si
LIGHTNING MVP System Lightning MVP System Control (HACCP+SSOP) Swab-uri pentru lichide si pentru Suprafete Accesorii ph Temperatura Condutivitate Monitorizare ATP Prin Bioluminescenta Cel mai complet si
Bioinformatics III Structural Bioinformatics and Genome Analysis Part Protein Secondary Structure Prediction. Sepp Hochreiter
 Bioinformatics III Structural Bioinformatics and Genome Analysis Part Protein Secondary Structure Prediction Institute of Bioinformatics Johannes Kepler University, Linz, Austria Chapter 4 Protein Secondary
Bioinformatics III Structural Bioinformatics and Genome Analysis Part Protein Secondary Structure Prediction Institute of Bioinformatics Johannes Kepler University, Linz, Austria Chapter 4 Protein Secondary
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
