Ubr3 E3 ligase regulates apoptosis by controlling the activity of DIAP1 in Drosophila
|
|
|
- Dale Lamb
- 6 years ago
- Views:
Transcription
1 (2014) 21, & 2014 Macmillan Publishers Limited All rights reserved /14 Ubr3 E3 ligase regulates apoptosis by controlling the activity of DIAP1 in Drosophila Q Huang 1,6, X Tang 6,2, G Wang 6,3, Y Fan 4, L Ray 2, A Bergmann 5, TY Belenkaya 2, X Ling 1, D Yan 1, Y Lin 1,XYe 1, W Shi 1, X Zhou 1,FLu 1, J Qu*,1 and X Lin*,1,2,3 Apoptosis has essential roles in a variety of cellular and developmental processes. Although the pathway is well studied, how the activities of individual components in the pathway are regulated is less understood. In Drosophila, a key component in apoptosis is Drosophila inhibitor of apoptosis protein 1 (DIAP1), which is required to prevent caspase activation. Here, we demonstrate that Drosophila CG42593 (ubr3), encoding the homolog of mammalian UBR3, has an essential role in regulating the apoptosis pathway. We show that loss of ubr3 activity causes caspase-dependent apoptosis in Drosophila eye and wing discs. Our genetic epistasis analyses show that the apoptosis induced by loss of ubr3 can be suppressed by loss of initiator caspase Drosophila Nedd2-like caspase (Dronc), or by ectopic expression of the apoptosis inhibitor p35, but cannot be rescued by overexpression of DIAP1. Importantly, we show that the activity of Ubr3 in the apoptosis pathway is not dependent on its Ring-domain, which is required for its E3 ligase activity. Furthermore, we find that through the UBR-box domain, Ubr3 physically interacts with the neoepitope of DIAP1 that is exposed after caspase-mediated cleavage. This interaction promotes the recruitment and ubiquitination of substrate caspases by DIAP1. Together, our data indicate that Ubr3 interacts with DIAP1 and positively regulates DIAP1 activity, possibly by maintaining its active conformation in the apoptosis pathway. (2014) 21, ; doi: /cdd ; published online 22 August 2014 Morphogenesis in multicellular organisms is a process with a balanced control of cell proliferation and cell death. To maintain this homeostasis, superfluous or unwanted cells are usually removed promptly via programmed cell death or apoptosis. 1,2 Compelling evidence has shown that dysregulation of apoptosis results in a variety of diseases, such as cancer, neurodegenerative disorders and autoimmune diseases. 3 6 The apoptotic machinery is conserved from invertebrates to vertebrates. Drosophila has been used as an excellent model to study apoptosis because of its advantages in genetic manipulation. A crucial step in apoptosis is the cascade activation of initiator and effector caspases that eventually causes cell death. Under normal circumstances, the activities of caspases are kept in check by a conserved family of anti-apoptotic proteins termed inhibitor of apoptosis proteins (IAPs). The Drosophila genome encodes four IAPs, including Drosophila inhibitor of apoptosis protein 1 (DIAP1), DIAP2, DBruce and Deterin Among these four proteins, DIAP1 is stringently required to prevent caspase activation. 11,12 Although the requirement of DIAP1 in the apoptosis pathway is well documented, it is unclear how the activity of DIAP1 is regulated during development. The covalent attachment of ubiquitin to proteins is a crucial regulatory mechanism in many developmental and physiological processes. 13 Ubiquitination is a catalytic cascade involving ubiquitin-activating (E1), ubiquitin-conjugating (E2) and ubiquitin-ligating (E3) enzymes. 14 The E3 proteins that specifically recognize a distinctive set of substrates for ubiquitination are an exceptionally large family. 15 The RING domain of DIAP1 is an E3 ligase that inactivates caspases mainly through ubiquitination. 16 Previous studies have shown that the anti-apoptotic activity of DIAP1 is negatively regulated by three pro-apoptotic proteins called Reaper, head involution defective (Hid) and Grim (RHG). 2,17 These proteins negatively regulate DIAP1 function through distinct mechanisms, either by disrupting interactions between DIAP1 and the initiator caspase Drosophila Nedd2-like caspase (Dronc), or by promoting the ubiquitination-dependent degradation of DIAP1. 18,19 In addition to regulation by RHG, DIAP1 has been considered a substrate of the N-end rule pathway. Ditzel et al. 20 first discovered that DIAP1 can be cleaved by effector caspases at its NH2 terminus, exposing a binding motif for UBR-box-containing E3 ligases and subsequently be degraded by the N-end rule-mediated degradation. Later, 1 School of Optometry and Ophthalmology and Eye Hospital, Wenzhou Medical University, Wenzhou, China; 2 Division of Developmental Biology, Cincinnati Children s Hospital Medical Center, Cincinnati, OH, USA; 3 State Key Laboratory of Biomembrane and Membrane Biotechnology, Institute of Zoology, Chinese Academy of Sciences, Beijing, China; 4 School of Biosciences, University of Birmingham, Birmingham, UK and 5 Department of Cancer Biology, University of Massachusetts Medical School, Worcester, MA, USA *Corresponding authors: X Lin, Division of Developmental Biology, Cincinnati Children s Hospital Medical Center, 3333 Burnet Avenue, Cincinnati 45229, OH, USA. Tel: ; Fax: ; xinhua.lin@cchmc.org or J Qu, School of Optometry and Ophthalmology and Eye Hospital, Wenzhou Medical University, 270 Xueyuan Road, Wenzhou , Zhejiang, China. Tel: ; jqu@wz.zj.cn 6 These authors contributed equally to this work. Abbreviations: IAPs, inhibitor of apoptosis proteins; DIAP1, Drosophila inhibitor of apoptosis protein 1; Dronc, Drosophila Nedd2-like caspase; En, engrailed; Ey, eyeless; Mirr, mirror; FLP, flippase; FRT, flippase recombination target; UAS, upstream activating sequence; GFP, green fluorescent protein; Hid, head involution defective; Hs, heat shock; TUNEL, terminal deoxynucleotidyl transferase dutp nick end labeling Received ; revised ; accepted ; Edited by E Baehrecke; published online
2 1962 Ubr3 regulates apoptosis Yokokura et al. 21 reported that the N-end rule pathway does not have a major role in the turnover of N-terminally truncated DIAP1. More recently, Ditzel et al. 22 reported that the UBRbinding motif of DIAP1 is essential for its anti-apoptotic function, indicating UBR family proteins regulate apoptosis via ways other than destroying DIAP1. Although these reports highlight the importance of UBR-box-containing E3 ligases in regulating DIAP1, it is currently unknown which UBR family member is involved in apoptosis. The mammalian genome encodes seven evolutionarily conserved members of UBR E3 ligases (UBR1 UBR7). 23,24 All of the UBR E3 ligases share a B70-amino-acid UBR-box and can function as N-recognins, which are involved in the N-end rule pathway of the ubiquitination system. 25 Among them, UBR1, UBR2 and UBR3 are subfamily members containing both a UBR-box and a Ring domain, which is required for its E3 ligase activity. 26 Data from mouse models indicate that UBR1 and UBR2 are involved in the regulation of apoptosis in spermatocytes, skeletal muscle and cardiovascular development with partial redundancy UBR3 was first characterized in mice as an E3 ligase involved in the regulation of olfactory and other sensory systems. 30 Recently, human UBR3 was also found to be required for genome stability by regulating the essential DNA repair protein APE1. 31 As UBR3 does not bind to the known N-end rule substrates of UBR1 and UBR2, 30 it is currently unknown whether UBR3 is involved in the apoptosis pathway. Here, we have generated Drosophila ubr3 mutant and characterized its role in development. Our data suggest that Ubr3 is involved in the apoptosis pathway by regulating the activity of DIAP1 during development. Results Disruption of ubr3 results in impaired eye and wing growth. The establishment of a genome-wide RNA interference (RNAi) library has facilitated genetic screening for genes affecting particular pathways or biological processes in Drosophila. To identify E3 ligases that are involved in regulating eye development, we carried out a systematic RNAi screen by selectively knocking down individual E3 ligases in the developing eye of Drosophila. One of the candidates identified from the screen was CG42953, which is the Drosophila homolog of UBR3 (ubr3). The Drosophila CG42593 mrna is predicted to encode a protein of 2219 amino acids with a theoretical molecular weight of 244 kda. Domain analysis and BlastP search shows that the amino-acid sequence of the UBR-box domain of CG42593 is 39% identical and 51% similar to the human UBR3 (NP_ ) (Figure 1a), indicating that CG42593 encodes the homolog of mammalian UBR3. As shown in Figures 2b and c, eyeless-gal4-induced ubr3 knock-down resulted in rough and smaller adult eyes. Similarly, knock-down of ubr3 by RNAi in the wing also impaired wing development (Supplementary Figure S1). To monitor the RNAi-mediated reduction of Ubr3, we generated an Ubr3-specific antibody and examined the protein levels by immunostaining. We observed that the expression of Ubr3 is greatly reduced in the dorsal compartment of the eye imaginal disc because of Mirror-Gal4-driven expression of ubr3 RNAi (Figure 2f). To further examine the role(s) of Ubr3 in eye development, we have generated a null allele of ubr3 by P-element-mediated imprecise excision (Figure 1b). Although the transcription of the gene CG2206, which lies within ubr3, is not affected by this allele (Supplementary Figure S1c), immunostaining indicates that expression of Ubr3 is completely absent in mutant clones (Figures 2g and g ). As the null allele of ubr3 is homozygous lethal, the adult eye phenotypes of this mutant were examined by virtue of the EGUF/hid method. 32 Consistent with the RNAi results, we observed strong eye defects including eye roughness and small eye size (Figure 2e). Loss of ubr3 in eye and wing discs induces apoptosis. To determine whether the aforementioned eye defects were caused by apoptosis, we used multiple methods to monitor the apoptosis in the eye and wing imaginal discs of third instar larvae upon ubr3 mutation and knockdown. We found that the levels of both activated caspases (cleaved caspase-3) and terminal deoxynucleotidyl transferase dutp nick end labeling (TUNEL) were significantly increased when ubr3 is knocked down by RNAi in the dorsal half of the eye disc (Figures 3a and b). We further performed mosaic clonal analysis using the ubr3 mutant in both eye and wing discs. Consistently, activation of caspase-3 was also observed in cells mutant for ubr3 (Figures 3c and d). To exclude the possibility that the eye phenotypes may also arise from defects in cell proliferation, we examined the mitosis status by staining with the mitotic marker phospho-histone H3 (PH3). As shown in Figures 3e-e and f, cell proliferation in ubr3 RNAi was not affected. Collectively, these data argue that Ubr3 is involved in the control of apoptosis rather than cell proliferation in Drosophila. Ubr3 acts upstream of Dronc, and downstream of or in parallel to DIAP1. A variety of evidence supports the view that DIAP1 is the primary suppressor of apoptosis in Drosophila. 11 Previous reports have shown that DIAP1 can be ubiquitinated by UBR family proteins (also called N-recognins) and degraded by proteasome. 33 To test this possibility, first we examined the DIAP1 protein levels in ubr3 mutant cells using a DIAP1-specific antibody (Figures 4a-a ), whose specificity is verified in Supplementary Figures S2a-a. The fact that protein levels of DIAP1 were slightly reduced only in some but not all ubr3 mutant cells excludes the possibility that Ubr3 targets DIAP1 for degradation. Although the change in DIAP1 levels may result from caspase activation in apoptotic cells, these data imply that Ubr3 do not directly control DIAP1 stability. Next, we checked the expression of Hid, one major pro-apoptotic proteins upstream of DIAP1. The increase in Hid protein levels is subtle if any in ubr3 mosaic clones (Figure 4b). Then, we asked whether the change in RHG complex is accounted for the apoptosis induced by ubr3 knockdown. We found that in both eye discs and wing discs (Figures 4c-d and Supplementary Figures S2c-e), ubr3 knockdown still induced significant cell death in clones of cells homozygous for H99, a deletion that removes rpr, hid and grim, arguing that ubr3 regulates apoptosis not mainly through RHG proteins.
3 Ubr3 regulates apoptosis 1963 Figure 1 Genomic and protein structures of Ubr3. (a) UBR-box domain of Ubr3 is highly conserved among different species. (b) Generation of a null allele by P-elementmediated imprecise excision. The dotted line indicates the deleted region, which includes the start codon and portion of N-terminal region We performed further genetic epistasis analysis to place ubr3 in the apoptosis pathway. In response to upstream death stimuli, the initiator caspase Dronc is auto-activated as a component of the apoptosome, and further activates effector caspases, including DrICE and DCP In this experiment, we knocked down ubr3 in a Dronc mutant background and found that the cell death induced by ubr3 RNAi was massively suppressed in the absence of Dronc (Figures 4e and f). This result argues that Ubr3 acts upstream of Dronc in the apoptosis pathway. Next, we asked whether overexpression of DIAP1 is able to suppress ubr3 RNAi-induced apoptosis. Although overexpression of DIAP1 can suppress GMR4rprinduced apoptosis (Supplementary Figure S3), it fails to block ubr3 RNAi-induced cell death in both eye discs and wing discs (Figures 4h-h and k-k ). In contrast, overexpression of baculovirus pancaspase inhibitor p35, which serves as an effective apoptosis inhibitor in Drosophila, 35 blocked the apoptosis in both cases (Figures 4i-i and l-l ). We also verified the apoptosis levels with acridine orange staining in eye discs (Supplementary Figure S4). Taken together, these results indicate that Ubr3 regulates apoptosis mainly in parallel to or downstream of DIAP1, but upstream of Dronc. The UBR-box domain rather than the Ring domain is essential for Ubr3 s anti-apoptotic function. Ubr3 contains two conserved domains, a UBR-box domain involved in protein protein interaction and a Ring domain required for its E3 ligase activity. 23,31 To further elucidate the molecular mechanism underlying Ubr3 s function in apoptosis, we dissected the role(s) of each domain by genetic rescue experiments using the MARCM system. 36 We generated transgenic flies expressing Gal4-inducible wild-type Ubr3 or two deletion mutants, Ubr3DUBR (UBR-box domain deleted) and Ubr3DRing (Ring domain deleted). Expression of Ubr3DRing or wild-type Ubr3 fully blocked the ectopic apoptosis in ubr3 mutant eye imaginal disc cells, as indicated by the absence of activated caspase-3 staining (Figures 5c-c and 5d-d ). In contrast, ectopic expression of Ubr3DUBR failed to suppress or only partially suppressed caspase-3 activation (Figures 5b-b ). Similar results were observed in wing discs (Supplementary Figure S5). In addition to caspase activation, we also examined the effect of different Ubr3 variants in restoring the eye size of ubr3 mutants. As shown before, by virtue of the EGUF/hid method, we were able to generate adult eyes composed of ubr3 mutant cells. Compared with the small and rough eyes of ubr3 mutants, expression of Ubr3DRing and wild-type Ubr3 both greatly restored the eye size (Figures 5f, Po0.05), while Ubr3DUBR showed little effect (Figures 5f, P40.05, n ¼ 8, one-way ANOVA with LSD post hoc). Representative pictures of each genotype are shown in Supplementary Figures S5. These data suggest that the UBR-box domain is required for Ubr3 s anti-apoptotic activity. However, the Ring domain is not essential, suggesting that Ubr3 may not act as an E3 ligase in this process. Among UBR family members in Drosophila,Ubr1(CG9086) and Ubr3 (CG42593) have similar domain organizations, as both contain a Ring domain and a UBR-box domain. Interestingly, when we overexpressed Ubr1 in the ubr3 RNAi background, no rescue of apoptosis was observed (Figures 5e-e ). Consistently, no significant difference was found in adult eye sizes between the two groups (Supplementary Figures S5 j l). These results suggest that Ubr1 and Ubr3 have distinct roles in Drosophila, despite the high similarities in protein structure.
4 Ubr3 regulates apoptosis 1964 Figure 2 Knockdown and knockout of ubr3 impaired the development of eye. All pictures are oriented anterior left, dorsal up. (a c) Compared with wild type (a), RNAimediated knockdown of ubr3 driven by Ey-Gal4 (b and c) induces growth defect in eyes. (d and e) Scanning electron microscope (SEM) analyses of adult eyes in ubr3 knockout mutant. Eye phenotypes were generated by virtue of EGUF/hid method.26 (d) FRT19A line was used as a control. (f) Ubr3 staining is greatly reduced in the dorsal compartment of the eye imaginal disc expressing ubr3 RNAi driven by Mirr-Gal4. (g-g ) Ubr3 protein is absent in ubr3 mutant clones. Genotypes: (b) w, UAS-ubr3 RNAi 1/EyGal4, (c) w,uas-ubr3 RNAi 2/Ey-Gal4, (d) FRT19A/yw,GMR-Hid, FRT19A; þ /Ey-Gal4, UAS-flp, (e) w,ubr31-frt19a/ yw,gmr-hid, FRT19A; þ /Ey-Gal4, UAS-flp, (f) w, þ / UAS-ubr3 RNAi 1; þ /Mirr-Gal4, (g) w, ubr31-frt19a /hsflp, Ubi-mRFP FRT19A Ubr3 binds to DIAP1 and promotes DIAP1-dependent caspases ubiquitination. As reported by Ditzel et al.,22 isolated UBR domain can recognize the neo-epitope of DIAP1 exposed after caspase-mediated cleavage. We have found that UBR3 regulates apoptosis in a UBR-boxdependent manner downstream of or in parallel to DIAP1. To further characterize the interaction between Ubr3 and DIAP1, we performed co-immunoprecipitation (co-ip) assays in cultured S2 cells. First, we confirmed that Ubr3 binds to the N-terminally cleaved form of DIAP1 (N-DIAP , the low band in lane 2), which exposes the binding motif for the UBR-box domain. However, Ubr3 fails to interact with fulllength DIAP1 (top band in lane2) (Figure 6a). Second, the physical interaction between Ubr3 and DIAP1 relies on the UBR-box domain and the neo-epitope of DIAP1. Either deletion of the UBR-box domain of Ubr3 or replacement of N21 to M21 in N-DIAP can destroy this interaction (Figure 6b). However, UBR3DRing can interact effectively with DIAP1, consistent with the in vivo findings that UBR3DRing is a functional protein, but UBR3DUBR is not (Figure 5). Similarly, we found that N-DIAP instead of M-DIAP can pull down endogenous Ubr3 (Supplementary Figure S6).
5 Ubr3 regulates apoptosis 1965 Figure 3 Loss of ubr3 activity induces apoptosis in eye imaginal discs. (a-b ) ubr3 was knocked-down by Mirr-Gal44RNAi in the dorsal compartment of eye disc. Dorsal is up as marked by GFP. Apoptosis was detected by activated caspase-3 (cleaved caspase-3 (Caspa3*)) staining (a-a ) and TUNEL assay (b-b ). (c-d ) Inubr3 mutant clones (GFP negative), Caspa3* is increased in eye disc (c) and wing disc (d). (e-e ) Eye discs labeled with mitosis marker PH3. Knockdown of ubr3 does not affect PH3 levels. (f) Quantification of PH3-positive cells. There is no statistical difference between the dorsal compartment (ubr3 RNAi) and ventral compartment (control) of the eye discs, as analyzed by paired t-test (P ¼ (two-tailed), n ¼ 9). Genotypes: (a, b and e) w; þ /UAS-ubr3 RNAi 1; þ /Mirr-Gal4 It has been shown that M-DIAP , which cannot be recognized by UBR family proteins, has reduced activities in catalyzing caspase ubiquitination and in inhibiting apoptosis. 22 Here, we further explored how the interaction of DIAP1 with Ubr3 affects DIAP1 s activities. As shown in Figure 6c, expression of N-DIAP in S2 cells readily ubiquitinated the downstream caspases Dronc and DrICE. Introduction of ectopic Ubr3 further enhanced N-DIAP induced caspase ubiquitination, while ubr3 knockdown significantly reduced DrICE ubiquitination (Figure 6c, the efficacy of Ubr3 RNAi is verified in Supplementary Figure S7), suggesting that Ubr3 regulates the anti-apoptotic activities of DIAP1. Although DIAP1 and Dronc/DrICE interact weakly with each other in normal cells, Ubr3 is found to be able to enhance their interaction possibly by forming a tertiary complex as shown by the co-ip results (Figure 6c). The interaction of Ubr3 with DIAP1 is important for the capability of Ubr3 in controlling DIAP1 s activities, as Ubr3 fails to affect M-DIAP s catalytic function in Dronc ubiquitination (Figure 6d). In support of this view, Ubr3DUBR, which fails to interact with DIAP1, has markedly reduced activity on DIAP1-induced DrICE ubiquitination (Supplementary Figure S8). Altogether, these results indicate that the presence of Ubr3 is required to maximize DIAP1 s E3 ligase activities on substrate caspases. Discussion In this study, we used the developing Drosophila eye and wing as in vivo models to analyze the function of ubr3 at the molecular and cellular levels. We showed that loss of ubr3 causes massive cell death in Drosophila. Our detailed genetic and molecular analyses led us to conclude that Ubr3 controls the apoptosis pathway by regulating DIAP1 activity. The Drosophila genome encodes six UBR-box-containing E3 ligases, and the functional details of each UBR protein remain elusive. Although DIAP1 has been shown as a substrate for N-end rule-mediated degradation and could interact with isolated UBR domains of multiple UBR family members, no single UBR protein has ever been identified in the regulation of DIAP1. 20,21 We for the first time characterized Ubr3, a highly conserved UBR protein, as an antiapoptotic factor, which controls the function rather than the stability of DIAP1. Interestingly, the role of Ubr3 in apoptosis pathway cannot be replaced by other UBR proteins such as Ubr1. Since N-end rule degradation pathway has been implicated in the regulation of DIAP1, it is possible that this process involves other UBR family members. More studies are needed to fully elucidate the role of UBR family proteins in the apoptosis pathway.
6 1966 Ubr3 regulates apoptosis
7 Ubr3 regulates apoptosis 1967 Our genetic epistasis experiments place Ubr3 in parallel to or downstream of DIAP1 in the apoptosis pathway. First, loss of RHG proteins fails to suppress cell death because of ubr3 knockdown, arguing that RHG complex is upstream of Ubr3 in the apoptotic pathway. Although Hid levels are slightly elevated in cells mutant for ubr3, it is probably a secondary effect because of the reduced activities of DIAP1. Interestingly, RHG proteins have been shown to be also ubiquitinated by DIAP1. 37 It is possible that ubr3 is also important for this activity of DIAP1. Second, Dronc mutation or ectopic expression of p35 can effectively suppress apoptosis induced by the loss of ubr3 activity. Third, overexpression of DIAP1 fails to inhibit loss-of-ubr3-induced apoptosis. Finally, we show that Ubr3 interacts with DIAP1 and promotes DIAP1- mediated caspase ubiquitination. On the basis of these data, we propose a model in which Ubr3 interacts with DIAP1 to regulate its activities in the apoptosis pathway. How does Ubr3 regulate DIAP1 s activity? Previously, Ditzel et al. 22 showed that the isolated UBR-box domains of UBR1, -3, -5, -6 and -7 can all bind to DIAP1. By using the M-DIAP mutant, which fails to interact with UBR proteins, they also showed that the UBR-DIAP1 interaction is required for DIAP1 s anti-apoptotic activity. However, the molecular nature of the involving UBR protein(s) was unclear. Our studies presented here resolved this issue by identifying Ubr3 as an essential protein required for DIAP1 activity. Moreover, we showed that overexpression of Ubr1, a homolog of Ubr3, fails to rescue the apoptosis defect in ubr3 mutant cells. This result shows that the activity of Ubr3 in the regulation of DIAP1 is highly specific, and strongly argues against the previous view that significant redundancy exists among UBR proteins in apoptotic regulation. It is important to mention that ectopic expression of an E3- defective Ubr3 can effectively suppress caspase-3 activation in ubr3 mutant cells, suggesting that Ubr3 does not act as an E3 ligase in regulating DIAP1 activity. Interestingly, we notice that Ring domain deletion renders Ubr3 more stable in cultured cells, implying a potential role of Ubr3 in its autoregulation through E3 ligase activity. Finally, we observed that Ubr3 forms a complex together with DIAP1 and caspases. Although Ubr3 regulates the ubiquitination levels of caspases, it also affects the interaction between caspases and their E3 ligase, DIAP1 (Figure 6d). Together, these data lead to a model in which Ubr3 promotes the interaction of DIAP1 with caspases, and therefore facilitates the subsequent ubiquitinating reaction (Figure 6e). Interestingly, recent data suggest that DIAP1 rests in an inactive conformation whose activation requires caspase-mediated cleavage. 22 As Ubr3 interacts with DIAP1 and caspases, the binding of Ubr3 to DIAP1 may also help to maintain DIAP1 in the active conformation, which is essential for its activity. Further biochemical and molecular experiments are needed to explore the detailed mechanisms underlying the regulation of DIAP1 s anti-apoptotic activity by Ubr3. Materials and Methods Drosophila stocks, genetics. Upstream activating sequence (UAS)-p35, UAS-DIAP1, actin-gal4, MS1096-Gal4, Eyeless-Gal4, mirror-gal4, yw FRT101, yw neofrt19a, yw Ubi-green fluorescent protein (GFP) FRT101, heat shockflippase (hs-flp), UAS-CD8::GFP, w hsflp, Tub-Gal80 FRT19A and GMR-Hid, yw FRT19A; eyeless-gal4, UAS-FLP lines were obtained from Bloomington Stock Center (Indiana University, Bloomington, IN, USA). MARCM clone tool lines yw, hsflp; tubgal4 UASGFP; FRT80B tubgal80 and yw; sp/cyo; FRT80B/TM6B were received from Dr. Lei Zhang (SIBCB, Shanghai, China). Dronc I29 is described previously. 34 H99 is a deletion of hid, rpr and grim. A null allele of ubr3 1 was generated by P-element-mediated imprecise excision from p (EP) EP1243, which deletes 1555 bp including the start codon and the first exon. ubr3 RNAi lines v (RNAi1) and v22901 (RNAi2) were obtained from Vienna Drosophila RNAi Center (VDRC), Vienna, Austria. Mosaic clones of ubr3 mutant were generated by the FLP-FRT method and induced in first/second-instar larvae, as described previously. 38 Flies were raised on standard cornmeal/yeast/molasses/agar media at 25 1C. Constructs and transgenic lines. Ubr3 cdna was generated by synthetic method from ESTs and exons, and details are available on request. puast-3flag-ubr3 was constructed by cloning the full-length ubr3 cdna into puast vector with XhoI and XbaI. Forward primer: 5 0 -ACGCTCGAGATGGACGA GGATGACAACCTG-3 0 ; reverse primer: 5 0 -ACGTCTAGACAAGAGATCCCGGTG AAACACCCATG-3 0. puast-3flag-ubr3dubr construct has a deletion of aa, and puast-3flag-ubr3dring construct has a deletion of aa. YFP-based DIAP1 construct is a modified version of the ubiquitin fusion construct described previously by Lévy et al. 39 Briefly, YFP was used as a reference protein to link to the N terminus of ubiquitin and the C terminus of ubiquitin was then linked to DIAP1. After cleavage of the tripartite fusion protein at the C terminus of ubiquitin, equimolar amounts of the unmodified DIAP1 and the reference protein will be produced. The YFP-Ubiquitin-DIAP1 construct was generated by overlapping PCR and cloned into puast vector with C-terminal V5 tag (details are available on request). Three different DIAP1 variants were generated by the same method: YFP-Ubiquitin-DIAP1-V5, YFP-Ubiquitin-N- DIAP V5 and YFP-Ubiquitin-M-DIAP V5, referred to as DIAP1(-V5), N-DIAP (-V5) and M-DIAP (-V5), respectively, in the text. DIAP1 (-V5) is a wild-type DIAP1 protein. N-DIAP (-V5) is a truncated DIAP1, from which the first 20 N-terminal amino acids are removed. In M-DIAP (-V5), the N 21 was replaced by M 21. RNAi-mediated knockdown of ubr3 in S2 cells was achieved by transfection of a short hairpin RNA transgene in pwalium20 knockdown vector. 40 A gene-specific 21-bp oligonucleotide (5 0 -GCACATTTGTGATGCACTACA-3 0 ) was selected and inserted into the vector. Eye size quantification and statistical analysis. All groups were crossed at the same time and raised under identical circumstances. All the pictures were taken on female flies at the same magnification, using AxioCam (Zeiss, Cambridge, UK). The areas of the eyes were measured by a colleague who is unaware of the genotypes, using Photoshop software (Adobe Systems Inc., Figure 4 Ubr3 functions in parallel to or downstream of DIAP1. (a-a ) Protein levels of DIAP1 are slightly reduced in some ubr3 mutant cells. (b-b ) The pro-apoptotic factor hid is slightly increased in cells mutant for ubr3.(c-d ) In the eye discs, loss of RHG proteins fails to block the cell death induced by Ubr3 knockdown. Ubr3 RNAi is expressed in control clones (c-c ) or clones homozygous for H99 (d-d ). All clones are positively marked by GFP. (e-f ) Ey4ubr3 RNAi-caused cell death in the eye discs is markedly suppressed by Dronc mutation. ubr3 RNAi is expressed throughout the eye discs in wild-type background (e-e ) ordronc homozygous mutant background (f-f ). (g-i ) Eye discs. (g-g ) Mirr4ubr3 RNAi induces apoptosis. (h-h ) Overexpression of wild-type DIAP1 fails to suppress apoptosis caused by Mirr4ubr3 RNAi. (i-i ) p35 overexpression is able to block the apoptosis caused by Mirr4ubr3 RNAi. (j-l ) In wing discs, cell death induced by En4ubr3 RNAi was examined when DIAP1 (k-k ) or p35 (l-l ) was co-expressed. Genotypes: (c) hsflp, tubgal4, UAS-GFP; UAS-RNAi1/ þ ; FRT80B/ tubgal80 FRT80B, (d) hsflp, tubgal4, UAS-GFP; RNAi1/ þ ; H99 FRT80B/ tubgal80 FRT80B, (e) w; UAS-ubr3 RNAi1/ Ey-Gal4,(f) w; Ey-Gal4/ubr3 RNAi1; Dronc I29,(g) w; þ /UAS-ubr3 RNAi 1; þ /Mirr-Gal4,(h) w; þ /UAS-ubr3 RNAi 1; UAS-DIAP1/ Mirr-Gal4, (i) w; þ /UAS-ubr3 RNAi 1; UAS-p35/Mirr-Gal4, (j) w; UAS-ubr3 RNAi 1/En-Gal4, (k) w; UAS-ubr3 RNAi 1/En-Gal4, þ / UAS-DIAP1, (l) w; UAS-ubr3 RNAi 1/En-Gal4, þ / UAS-p35
8 1968 Ubr3 regulates apoptosis Figure 5 Genetic rescue of apoptosis in ubr3 mutant by overexpression of different Ubr3 constructs and Ubr1. (a-d ) MARCM clone analysis (GFP as positive labeling marker). (a-a ) Caspase-3 is activated in ubr3 mutant clones. (b-d ) Caspase-3 activation is rescued by expression of Ubr3DRing or Ubr3 (WT). Ubr3DUBR expression partially suppresses caspase-3 activation. (e-e ) Overexpression of Ubr1 fails to inhibit caspase-3 activation in ubr3 deficient cells. (f) Statistical analysis of the sizes of ubr3 mutant eyes rescued by expression of different Ubr3 variants (one-way ANOVA with LSD post hoc, *denotes Po0.05, # denotes P40.05). Ubr3DRing and Ubr3 show rescuing effects, but not Ubr3DUBR. Genotypes: (a) ubr3 1 -FRT19A/w, hsflp, Tub-Gal80 FRT19A; þ /Act-Gal4,UAS-GFP, (b) ubr3 1 -FRT19A/w, hsflp, Tub-Gal80 FRT19A; þ /Act-Gal4, UAS-GFP; þ /UAS-ubr3DUBR, (c) ubr3 1 -FRT19A/w, hsflp, Tub-Gal80 FRT19A; þ /Act-Gal4, UAS-GFP; þ /UAS-ubr3DRing, (d) ubr3 1 -FRT19A/w, hsflp, Tub-Gal80 FRT19A; þ /Act-Gal4, UAS-GFP; þ /UAS-ubr3, (e) w; þ /UAS-ubr3 RNAi 1; UAS-ubr1/Mirr-Gal4
9 Ubr3 regulates apoptosis 1969 Figure 6 DIAP1 binds to Ubr3 and promotes ubiquitination of DrICE and Dronc. (a) Co-IP assays with Ubr3 and DIAP1. S2 cells were co-transfected with V5-tagged wildtype DIAP1 or DIAP and Flag-tagged Ubr3. Co-precipitated proteins were analyzed by immunoblotting (IB) with the indicated antibodies. The wild-type DIAP1 exhibits two bands, the full length (top band) and the N-terminally cleaved one (low band). The interaction of Ubr3 with the cleaved DIAP1 (DIAP ) but not the full-length DIAP1 can be detected. (b) Co-IPs were performed as in (a) with N- and M-DIAP and the Ubr3 variants. (c) Effects of Ubr3 co-expression or knockdown on DIAP1-mediated caspase ubiquitination. S2 cells were co-transfected with myc-tagged Dronc/DrICE together with N-DIAP and constructs encoding Flag-tagged Ubr3 or Ubr3 shrna. Half of the cells were lysed under denaturing conditions using hot lysis protocol and ubiquitinated caspases were pulled down by myc affinity gel followed by immunoblotting with anti-ubiquitin antibody. The other half of cells were processed for co-ip assays as in (a). (d) Ubr3 fails to affect M-DIAP s catalytic function in Dronc ubiquitination. Experiments were performed similar to (c). (e) Model of Ubr3 s function in apoptosis. Ubr3 helps block apoptosis mainly through its interaction with DIAP1. This interaction promotes the binding of downstream caspases to DIAP1 and facilitates subsequent ubiquitination reactions San Jose, CA, USA). Data were analyzed with SPSS15.0 (IBM Corp., Chicago, IL, USA), using one-way ANOVA with LSD post hoc tests. All data were presented as means±s.d., and Po0.05 was considered statistically significant. Apoptosis assays. For TUNEL assay, dissected larval and pupal tissues were fixed for 20 min at RT in 4% formaldehyde in PBST, washed in PBST (0.3% Triton X-100) and permeabilized by incubation in 100 mm Na citrate/0.1% Triton X-100 at 65 1C for 30 min. Samples were rinsed in PBST and TUNEL was performed using a kit from Roche (Indianapolis, IN, USA). Tissues were mounted for confocal analysis (Zeiss LSM780). Immunostaining. Dissection, fixation and immunostaining of imaginal discs were performed as described previously. 41 Guinea pig anti-ubr3 primary antibody was produced against amino acids Guinea pig anti-ubr1 primary antibody was targeted against amino acids Primary antibodies used for the immunostainings were rabbit anti-cleaved caspase-3 (Cell Signaling Technology, Danvers, MA, USA), Mouse anti-ph3 (Abcam, Cambridge, MA, USA), rabbit anti-hid (d-300) (Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti Flag (Sigma, St. Louis, MO, USA) and rabbit anti-gfp Alexa Fluor 488 (Molecular Probes, Eugene, OR, USA). Mouse anti-diap1 (1:5) was generously provided by Dr. Bruce A Hay (California Institute of Technology, Pasadena, CA, USA). The primary antibodies were detected by fluorescent-conjugated secondary antibodies from Jackson ImmunoResearch (West Grove, PA, USA). Immunoprecipitation and western blotting. Drosophila S2 cells were maintained at 25 1C in Hyclone SFX-Insect cell culture medium (SH , Thermo Scientific, Waltham, MA, USA). For co-ip experiments, Drosophila S2 cells were transfected in 60 mm dishes with 2 mg of total DNA, using Effectene (Qiagen, Hilden, Germany). Forty-eight hours after transfection, cells were harvested and lysed in 1% Trition X-100 lysis buffer: 1% Triton X-100, 20 mm Tris (ph 7.5), 1 mm EDTA, 150 mm NaCl, protease inhibitor cocktail (Roche, added right before use). Co-IP and western blot experiments were performed as described. 38 For ubiquitination assays, transfected cells were treated with 50 mm MG-132 (Sigma) for 6 h before harvest. Forty-eight hours after transfection, cells were harvested and lysed by hot lysis protocol. Generally, cells were boiled at 100 1C in hot lysis buffer I (1% SDS, 50 mm NaF and 1 mm EDTA) followed by extensive shearing and mixing. After centrifugation and fivefolds dilution with hot lysis buffer II (25 mm Tris-HCl, ph 7.5, 1.25% Triton X-100, 125 mm NaCl and 50 mm NaF, protease inhibitors and a final concentration of 50 mm MG132 were added just before use), the supernatant was collected as cell lysate. The following antibody-bound beads were used for immunoprecipitation: EZview Red Anti-FLAG M2 Affinity Gel (Sigma), EZview Red Anti-c-Myc Affinity Gel (Sigma), Anti-V5 Agarose Affinity Gel (Sigma). The primary antibodies used for western blotting were rabbit anti-v5 (Sigma), mouse anti-v5 (Invitrogen, Carlsbad, CA, USA), mouse anti-ub (P4D1, Santa Cruz Biotechnology), mouse anti-flag (Sigma) and mouse anti-myc (Sigma). Conflict of Interest The authors declare no conflict of interest. Acknowledgements. We thank Dr. Bruce A Hay (Caltech, USA) for providing the anti-diap1 antibody and Dr. Meier for sending us various tub4diap1 transgenic lines and the TRiP at Harvard Medical School (NIH/NIGMS R01- GM084947) for providing the pvalium20 vector. Many lines are obtained from the Bloomington Stock Center and the Vienna Drosophila RNAi Center. This work is supported by grants the National Basic Research Program of China
10 1970 Ubr3 regulates apoptosis (2011CB and 2011CB943902), Research Foundation for Advanced Talents of Wenzhou Medical University (QTJ08012) and Wenzhou Medical University research grant (XNK07005), National Natural Science Foundation of China ( ), NIH grants (2R01 GM and 1R01GM087517) and Strategic Priority Research Program of the Chinese Academy of Sciences Grant (XDA ). YF is supported by Marie Curie Career Integration Grant from the EU FP7 Program and the Birmingham Fellowship, University of Birmingham, UK. AB is supported by Grant R01 GM from the National Institute of General Medical Sciences (NIGMS). 1. Vaux DL, Korsmeyer SJ. Cell death in development. Cell 1999; 96: Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell 2011; 147: Baehrecke EH. How death shapes life during development. Nat Rev Mol Cell Biol 2002; 3: Vaux DL, Flavell RA. Apoptosis genes and autoimmunity. Curr Opin Immunol 2000; 12: Yuan J, Yankner BA. Apoptosis in the nervous system. Nature 2000; 407: Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100: Kaiser WJ, Vucic D, Miller LK. The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drice. FEBS Lett 1998; 440: Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ et al. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem 2007; 282: Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol 2007; 5: e Jones G, Jones D, Zhou L, Steller H, Deterin Chu Y. a new inhibitor of apoptosis from Drosophila melanogaster. J Biol Chem 2000; 275: Srinivasula SM, Ashwell JD. IAPs: what s in a name? Mol Cell 2008; 30: Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 2002; 277: Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature 2009; 458: Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 2006; 22: Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 2008; 9: Bergmann A. The role of ubiquitylation for the control of cell death in Drosophila. Cell Death Differ 2010; 17: Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ 2008; 15: Varshavsky A. The N-end rule and regulation of apoptosis. Nat Cell Biol 2003; 5: Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 2002; 4: Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol 2003; 5: Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P et al. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem 2004; 279: Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT et al. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell 2008; 32: Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol 2005; 25: Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci 2007; 32: Sriram SM, Kim BY, Kwon YT. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol 2011; 12: Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 2000; 288: Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol Cell Biol 2001; 21: Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol 2003; 23: An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, Kwon YT. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci USA 2006; 103: Tasaki T, Sohr R, Xia Z, Hellweg R, Hortnagl H, Varshavsky A et al. Biochemical and genetic studies of UBR3, a ubiquitin ligase with a function in olfactory and other sensory systems. J Biol Chem 2007; 282: Meisenberg C, Tait PS, Dianova II, Wright K, Edelmann MJ, Ternette N et al. Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability. Nucleic Acids Res 2012; 40: Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 1999; 152: Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ 2007; 14: Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development 2005; 132: Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development 1994; 120: Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 2001; 24: Olson MR, Holley CL, Yoo SJ, Huh JR, Hay BA, Kornbluth S. Reaper is regulated by IAPmediated ubiquitination. J Biol Chem 2003; 278: Belenkaya TY, Han C, Standley HJ, Lin X, Houston DW, Heasman J. pygopus Encodes a nuclear protein essential for wingless/wnt signaling. Development 2002; 129: Levy F, Johnsson N, Rumenapf T, Varshavsky A. Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci USA 1996; 93: Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D et al. A genome-scale shrna resource for transgenic RNAi in Drosophila. Nat Methods 2011; 8: Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV et al. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-golgi network. Dev Cell 2008; 14: Supplementary Information accompanies this paper on website (
Drosophila Apoptosis and the Regulation of the Caspase Cascade
 Drosophila Apoptosis and the Regulation of the Caspase Cascade Kate Stafford March 18, 2005 Abstract The caspase cascade in Drosophila is controlled primarily by DIAP1 (Drosophila inhibitor of apoptosis),
Drosophila Apoptosis and the Regulation of the Caspase Cascade Kate Stafford March 18, 2005 Abstract The caspase cascade in Drosophila is controlled primarily by DIAP1 (Drosophila inhibitor of apoptosis),
Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation
 University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 9-1-2011 Drosophila IAP1-mediated ubiquitylation controls
University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 9-1-2011 Drosophila IAP1-mediated ubiquitylation controls
Chapter 4: A Deficiency Screen to Isolate Novel Regulators of DIAP1
 70 Chapter 4: A Deficiency Screen to Isolate Novel Regulators of DIAP1 4.1 Abstract DIAP1, a ubiquitin E3 ligase, is the key Drosophila regulator of the family of cysteine proteases known as caspases.
70 Chapter 4: A Deficiency Screen to Isolate Novel Regulators of DIAP1 4.1 Abstract DIAP1, a ubiquitin E3 ligase, is the key Drosophila regulator of the family of cysteine proteases known as caspases.
S Phase Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
 S Phase Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues Jean M. Davidson 1, Robert J. Duronio 1,2,3 * 1 Department of Biology, The University of North Carolina at Chapel Hill, Chapel
S Phase Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues Jean M. Davidson 1, Robert J. Duronio 1,2,3 * 1 Department of Biology, The University of North Carolina at Chapel Hill, Chapel
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
Supplementary Information
 Supplementary Information Supplementary Figure 1. JAK/STAT in early wing development (a-f) Wing primordia of second instar larvae of the indicated genotypes labeled to visualize expression of upd mrna
Supplementary Information Supplementary Figure 1. JAK/STAT in early wing development (a-f) Wing primordia of second instar larvae of the indicated genotypes labeled to visualize expression of upd mrna
Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc
 OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
Apoptosis in Mammalian Cells
 Apoptosis in Mammalian Cells 7.16 2-10-05 Apoptosis is an important factor in many human diseases Cancer malignant cells evade death by suppressing apoptosis (too little apoptosis) Stroke damaged neurons
Apoptosis in Mammalian Cells 7.16 2-10-05 Apoptosis is an important factor in many human diseases Cancer malignant cells evade death by suppressing apoptosis (too little apoptosis) Stroke damaged neurons
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
TNFα 18hr. Control. CHX 18hr. TNFα+ CHX 18hr. TNFα: 18 18hr (KDa) PARP. Cleaved. Cleaved. Cleaved. Caspase3. Pellino3 shrna. Control shrna.
 Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
purpose of this Chapter is to highlight some problems that will likely provide new
 119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
Caspase-Dependent Cell Death in Drosophila
 Annu. Rev. Cell Dev. Biol. 2006. 22:623 50 First published online as a Review in Advance on July 14, 2006 The Annual Review of Cell and Developmental Biology is online at http://cellbio.annualreviews.org
Annu. Rev. Cell Dev. Biol. 2006. 22:623 50 First published online as a Review in Advance on July 14, 2006 The Annual Review of Cell and Developmental Biology is online at http://cellbio.annualreviews.org
Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery Introduction UNCORRECTED PROOF
 (2006) 00, 1 12 & 2006 Nature Publishing Group All rights reserved 1350-9047/06 $30.00 www.nature.com/cdd Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery
(2006) 00, 1 12 & 2006 Nature Publishing Group All rights reserved 1350-9047/06 $30.00 www.nature.com/cdd Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Interactions of DNR1 with the apoptotic machinery of Drosophila melanogaster
 Research Article 1189 Interactions of DNR1 with the apoptotic machinery of Drosophila melanogaster David A. Primrose, Sidharth Chaudhry, A. George D. Johnson, Adam Hrdlicka, Anja Schindler, Dave Tran and
Research Article 1189 Interactions of DNR1 with the apoptotic machinery of Drosophila melanogaster David A. Primrose, Sidharth Chaudhry, A. George D. Johnson, Adam Hrdlicka, Anja Schindler, Dave Tran and
Mechanism of Dronc activation in Drosophila cells
 Research Article 5035 Mechanism of Dronc activation in Drosophila cells Israel Muro*, Kristin Monser* and Rollie J. Clem Molecular, Cellular, and Developmental Biology Program, Division of Biology, Ackert
Research Article 5035 Mechanism of Dronc activation in Drosophila cells Israel Muro*, Kristin Monser* and Rollie J. Clem Molecular, Cellular, and Developmental Biology Program, Division of Biology, Ackert
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Cell Death & Trophic Factors II. Steven McLoon Department of Neuroscience University of Minnesota
 Cell Death & Trophic Factors II Steven McLoon Department of Neuroscience University of Minnesota 1 Remember? Neurotrophins are cell survival factors that neurons get from their target cells! There is a
Cell Death & Trophic Factors II Steven McLoon Department of Neuroscience University of Minnesota 1 Remember? Neurotrophins are cell survival factors that neurons get from their target cells! There is a
This is an open access publisher version of an article that appears in:
 This is an open access publisher version of an article that appears in: JOURNAL OF CELL BIOLOGY The internet address for this paper is: https://publications.icr.ac.uk/5622/ Published text: PS Ribeiro,
This is an open access publisher version of an article that appears in: JOURNAL OF CELL BIOLOGY The internet address for this paper is: https://publications.icr.ac.uk/5622/ Published text: PS Ribeiro,
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila
 Research article 5591 Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila Ainhoa Pérez-Garijo, Francisco A. Martín and Ginés Morata* Centro de Biología
Research article 5591 Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila Ainhoa Pérez-Garijo, Francisco A. Martín and Ginés Morata* Centro de Biología
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice Supplementary Figure 2.
 Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Role of Mitochondrial Remodeling in Programmed Cell Death in
 Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
The Caspase System: a potential role in muscle proteolysis and meat quality? Tim Parr
 The Caspase System: a potential role in muscle proteolysis and meat quality? Tim Parr Caroline Kemp, Ron Bardsley,, Peter Buttery Division of Nutritional Sciences, School of Biosciences, University of
The Caspase System: a potential role in muscle proteolysis and meat quality? Tim Parr Caroline Kemp, Ron Bardsley,, Peter Buttery Division of Nutritional Sciences, School of Biosciences, University of
Characterization of the apoptotic functions of the HID hmolog isolated from Megaselia scalaris
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2013 Characterization of the apoptotic
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2013 Characterization of the apoptotic
Characterization of head involution defective (hid) as a pro-apoptotic gene in Megasalia scalaris
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2015 Characterization of head
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2015 Characterization of head
Types of biological networks. I. Intra-cellurar networks
 Types of biological networks I. Intra-cellurar networks 1 Some intra-cellular networks: 1. Metabolic networks 2. Transcriptional regulation networks 3. Cell signalling networks 4. Protein-protein interaction
Types of biological networks I. Intra-cellurar networks 1 Some intra-cellular networks: 1. Metabolic networks 2. Transcriptional regulation networks 3. Cell signalling networks 4. Protein-protein interaction
Regulation of the Drosophila Initiator Caspase Dronc through Ubiquitylation
 University of Massachusetts Medical School escholarship@umms GSBS Dissertations and Theses Graduate School of Biomedical Sciences 1-17-2017 Regulation of the Drosophila Initiator Caspase Dronc through
University of Massachusetts Medical School escholarship@umms GSBS Dissertations and Theses Graduate School of Biomedical Sciences 1-17-2017 Regulation of the Drosophila Initiator Caspase Dronc through
Supporting Information
 Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila
 Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
Segment boundary formation in Drosophila embryos
 Segment boundary formation in Drosophila embryos Development 130, August 2003 Camilla W. Larsen, Elizabeth Hirst, Cyrille Alexandre and Jean Paul Vincent 1. Introduction: - Segment boundary formation:
Segment boundary formation in Drosophila embryos Development 130, August 2003 Camilla W. Larsen, Elizabeth Hirst, Cyrille Alexandre and Jean Paul Vincent 1. Introduction: - Segment boundary formation:
Drosophila Bruce Can Potently Suppress Rpr- and Grim-Dependent but Not Hid-Dependent Cell Death
 Current Biology, Vol. 12, 1164 1168, July 9, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)00935-1 Drosophila Bruce Can Potently Suppress Rpr- and Grim-Dependent but Not Hid-Dependent
Current Biology, Vol. 12, 1164 1168, July 9, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)00935-1 Drosophila Bruce Can Potently Suppress Rpr- and Grim-Dependent but Not Hid-Dependent
RNA Synthesis and Processing
 RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1
 Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Information
 Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
FSC-W FSC-H CD4 CD62-L
 Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis Somasekar Seshagiri* and Lois K. Miller*
 Research Paper 455 Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis Somasekar Seshagiri* and Lois K. Miller* Background: Programmed cell death or apoptosis is a key
Research Paper 455 Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis Somasekar Seshagiri* and Lois K. Miller* Background: Programmed cell death or apoptosis is a key
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
The geneticist s questions. Deleting yeast genes. Functional genomics. From Wikipedia, the free encyclopedia
 From Wikipedia, the free encyclopedia Functional genomics..is a field of molecular biology that attempts to make use of the vast wealth of data produced by genomic projects (such as genome sequencing projects)
From Wikipedia, the free encyclopedia Functional genomics..is a field of molecular biology that attempts to make use of the vast wealth of data produced by genomic projects (such as genome sequencing projects)
Chapter 3: Novel Regulators of the C. elegans Caspase CED-3
 53 Chapter 3: Novel Regulators of the C. elegans Caspase CED-3 3.1 Abstract The hallmark of apoptosis is the presence of activated caspases, a conserved family of cysteine proteases. Proper regulation
53 Chapter 3: Novel Regulators of the C. elegans Caspase CED-3 3.1 Abstract The hallmark of apoptosis is the presence of activated caspases, a conserved family of cysteine proteases. Proper regulation
L œil de la Drosophile pour l étude du développement et de l apoptose
 L œil de la Drosophile pour l étude du développement et de l apoptose Pr Bertrand Mollereau Apoptosis and Neurogenetic LBMC UMR5239 ENS Lyon http://www.ens-lyon.fr/lbmc/apodroso/ Drosophila is a powerful
L œil de la Drosophile pour l étude du développement et de l apoptose Pr Bertrand Mollereau Apoptosis and Neurogenetic LBMC UMR5239 ENS Lyon http://www.ens-lyon.fr/lbmc/apodroso/ Drosophila is a powerful
Introduction. Gene expression is the combined process of :
 1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
NOPO modulates Egr-induced JNK-independent cell death in Drosophila
 ORIGINAL ARTICLE Cell Research (2012) 22:425-431. 2012 IBCB, SIBS, CAS All rights reserved 1001-0602/12 $ 32.00 www.nature.com/cr npg NOPO modulates Egr-induced JNK-independent cell death in Drosophila
ORIGINAL ARTICLE Cell Research (2012) 22:425-431. 2012 IBCB, SIBS, CAS All rights reserved 1001-0602/12 $ 32.00 www.nature.com/cr npg NOPO modulates Egr-induced JNK-independent cell death in Drosophila
A complementation test would be done by crossing the haploid strains and scoring the phenotype in the diploids.
 Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
TO DIE OR TO DIFFERENTIATE: APOPTOTIC AND NON-APOPTOTIC ROLES OF DEATH MOLECULES IN DROSOPHILA MELANOGASTER
 TO DIE OR TO DIFFERENTIATE: APOPTOTIC AND NON-APOPTOTIC ROLES OF DEATH MOLECULES IN DROSOPHILA MELANOGASTER Thesis by Jun Ryul Huh In Partial Fulfillment of the Requirements for the Degree of Doctor of
TO DIE OR TO DIFFERENTIATE: APOPTOTIC AND NON-APOPTOTIC ROLES OF DEATH MOLECULES IN DROSOPHILA MELANOGASTER Thesis by Jun Ryul Huh In Partial Fulfillment of the Requirements for the Degree of Doctor of
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
Bypass and interaction suppressors; pathway analysis
 Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
Autophagy-independent function of Atg1 for apoptosis-induced compensatory proliferation
 University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 8-19-2016 Autophagy-independent function of Atg1 for apoptosis-induced
University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 8-19-2016 Autophagy-independent function of Atg1 for apoptosis-induced
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression Differential gene expression Every somatic cell in an individual organism contains the same genetic information and replicated from the same original fertilized
Chapter 18 Regulation of Gene Expression Differential gene expression Every somatic cell in an individual organism contains the same genetic information and replicated from the same original fertilized
Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain
 Development 127, 367-379 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV5342 367 Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain Wei Du
Development 127, 367-379 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV5342 367 Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain Wei Du
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Eukaryotic Gene Expression
 Eukaryotic Gene Expression Lectures 22-23 Several Features Distinguish Eukaryotic Processes From Mechanisms in Bacteria 123 Eukaryotic Gene Expression Several Features Distinguish Eukaryotic Processes
Eukaryotic Gene Expression Lectures 22-23 Several Features Distinguish Eukaryotic Processes From Mechanisms in Bacteria 123 Eukaryotic Gene Expression Several Features Distinguish Eukaryotic Processes
The geneticist s questions
 The geneticist s questions a) What is consequence of reduced gene function? 1) gene knockout (deletion, RNAi) b) What is the consequence of increased gene function? 2) gene overexpression c) What does
The geneticist s questions a) What is consequence of reduced gene function? 1) gene knockout (deletion, RNAi) b) What is the consequence of increased gene function? 2) gene overexpression c) What does
Mitochondrial fusion is regulated by Reaper to modulate Drosophila programmed cell death
 (2011) 18, 1640 1650 & 2011 Macmillan Publishers Limited All rights reserved 1350-9047/11 www.nature.com/cdd Mitochondrial fusion is regulated by Reaper to modulate Drosophila programmed cell death M Thomenius
(2011) 18, 1640 1650 & 2011 Macmillan Publishers Limited All rights reserved 1350-9047/11 www.nature.com/cdd Mitochondrial fusion is regulated by Reaper to modulate Drosophila programmed cell death M Thomenius
Developmental Biology
 Developmental Biology 358 (2011) 213 223 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Cks85A and Skp2 interact to maintain diploidy
Developmental Biology 358 (2011) 213 223 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Cks85A and Skp2 interact to maintain diploidy
Full-length GlpG sequence was generated by PCR from E. coli genomic DNA. (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
 Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
Principles of Genetics
 Principles of Genetics Snustad, D ISBN-13: 9780470903599 Table of Contents C H A P T E R 1 The Science of Genetics 1 An Invitation 2 Three Great Milestones in Genetics 2 DNA as the Genetic Material 6 Genetics
Principles of Genetics Snustad, D ISBN-13: 9780470903599 Table of Contents C H A P T E R 1 The Science of Genetics 1 An Invitation 2 Three Great Milestones in Genetics 2 DNA as the Genetic Material 6 Genetics
Zool 3200: Cell Biology Exam 5 4/27/15
 Name: Trask Zool 3200: Cell Biology Exam 5 4/27/15 Answer each of the following short answer questions in the space provided, giving explanations when asked to do so. Circle the correct answer or answers
Name: Trask Zool 3200: Cell Biology Exam 5 4/27/15 Answer each of the following short answer questions in the space provided, giving explanations when asked to do so. Circle the correct answer or answers
 DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering
 Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Bio/Life: Cell Biology
 Bio/Life: Cell Biology 1a The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism's cells. As a basis for understanding
Bio/Life: Cell Biology 1a The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism's cells. As a basis for understanding
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
Regulation of gene expression. Premedical - Biology
 Regulation of gene expression Premedical - Biology Regulation of gene expression in prokaryotic cell Operon units system of negative feedback positive and negative regulation in eukaryotic cell - at any
Regulation of gene expression Premedical - Biology Regulation of gene expression in prokaryotic cell Operon units system of negative feedback positive and negative regulation in eukaryotic cell - at any
A poptosis is an evolutionarily conserved process through
 Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms Soon Ji Yoo*, Jun R. Huh*, Israel Muro, Hong Yu*, Lijuan Wang*, Susan L. Wang*, R. M. Renny Feldman*, Rollie J. Clem, H.-Arno
Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms Soon Ji Yoo*, Jun R. Huh*, Israel Muro, Hong Yu*, Lijuan Wang*, Susan L. Wang*, R. M. Renny Feldman*, Rollie J. Clem, H.-Arno
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX.
 Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A)
 Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Midterm 1. Average score: 74.4 Median score: 77
 Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON
 PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
09/30/2017. Kyu-Sun Lee, Ph.D. Metabolism & Neurophysiology Research Group Hazard Monitoring BNT Res Center
 09/30/2017 Kyu-Sun Lee, Ph.D. Metabolism & Neurophysiology Research Group Hazard Monitoring BNT Res Center Conflict of interest disclosure None Committee of Scientific Affairs Committee of Scientific Affairs
09/30/2017 Kyu-Sun Lee, Ph.D. Metabolism & Neurophysiology Research Group Hazard Monitoring BNT Res Center Conflict of interest disclosure None Committee of Scientific Affairs Committee of Scientific Affairs
The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila
 2125 The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila Dongbin Xu 1, *, Ying Li 1, *, Michael Arcaro 1, Melinda Lackey 1 and Andreas Bergmann 1,
2125 The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila Dongbin Xu 1, *, Ying Li 1, *, Michael Arcaro 1, Melinda Lackey 1 and Andreas Bergmann 1,
Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Tuesday, December 27, 16
 Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Enduring understanding 3.B: Expression of genetic information involves cellular and molecular
Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Enduring understanding 3.B: Expression of genetic information involves cellular and molecular
Regulation of Phosphate Homeostasis by microrna in Plants
 Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
CASPASES AND CASPASE REGULATORS IN LEPIDOPTERA AND DIPTERA WILLIAM BARTON BRYANT. B.S., Middle Tennessee State University, 2002
 CASPASES AND CASPASE REGULATORS IN LEPIDOPTERA AND DIPTERA by WILLIAM BARTON BRYANT B.S., Middle Tennessee State University, 2002 AN ABSTRACT OF A DISSERTATION submitted in partial fulfillment of the requirements
CASPASES AND CASPASE REGULATORS IN LEPIDOPTERA AND DIPTERA by WILLIAM BARTON BRYANT B.S., Middle Tennessee State University, 2002 AN ABSTRACT OF A DISSERTATION submitted in partial fulfillment of the requirements
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY METHODS Mef2 primary screen. RNAi hairpins from the VDRC collection were crossed to Mef2-GAL4 at 27 C. After 2 weeks lethality rate and stage was scored, and if possible 20-30 males containing
SUPPLEMENTARY METHODS Mef2 primary screen. RNAi hairpins from the VDRC collection were crossed to Mef2-GAL4 at 27 C. After 2 weeks lethality rate and stage was scored, and if possible 20-30 males containing
Chang 1. Characterization of the Drosophila Ortholog of the Mammalian Anti-apoptotic Protein Aven
 Chang 1 Characterization of the Drosophila Ortholog of the Mammalian Anti-apoptotic Protein Aven Joy Chang Georgetown University Senior Digital Thesis May 1, 2006 Chang 2 Table of Contents BACKGROUND 4
Chang 1 Characterization of the Drosophila Ortholog of the Mammalian Anti-apoptotic Protein Aven Joy Chang Georgetown University Senior Digital Thesis May 1, 2006 Chang 2 Table of Contents BACKGROUND 4
Developmental genetics: finding the genes that regulate development
 Developmental Biology BY1101 P. Murphy Lecture 9 Developmental genetics: finding the genes that regulate development Introduction The application of genetic analysis and DNA technology to the study of
Developmental Biology BY1101 P. Murphy Lecture 9 Developmental genetics: finding the genes that regulate development Introduction The application of genetic analysis and DNA technology to the study of
Supplementary Figure 1.
 Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
7.06 Spring 2004 PS 6 KEY 1 of 14
 7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus:
 m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
2. Der Dissertation zugrunde liegende Publikationen und Manuskripte. 2.1 Fine scale mapping in the sex locus region of the honey bee (Apis mellifera)
 2. Der Dissertation zugrunde liegende Publikationen und Manuskripte 2.1 Fine scale mapping in the sex locus region of the honey bee (Apis mellifera) M. Hasselmann 1, M. K. Fondrk², R. E. Page Jr.² und
2. Der Dissertation zugrunde liegende Publikationen und Manuskripte 2.1 Fine scale mapping in the sex locus region of the honey bee (Apis mellifera) M. Hasselmann 1, M. K. Fondrk², R. E. Page Jr.² und
Supplemental Materials Molecular Biology of the Cell
 Supplemental Materials Molecular iology of the Cell Figure S1 Krüger et al. Arabidopsis Plasmodium H. sapiens* 1) Xenopus* 1) Drosophila C.elegans S.cerevisae S.pombe L.major T.cruzi T.brucei DCP5 CITH
Supplemental Materials Molecular iology of the Cell Figure S1 Krüger et al. Arabidopsis Plasmodium H. sapiens* 1) Xenopus* 1) Drosophila C.elegans S.cerevisae S.pombe L.major T.cruzi T.brucei DCP5 CITH
The Caenorhabditis elegans CED-9 protein does not directly inhibit the caspase CED-3, in vitro nor in yeast
 (2004) 11, 1309 1316 & 2004 Nature Publishing Group All rights reserved 1350-9047/04 $30.00 www.nature.com/cdd The Caenorhabditis elegans CED-9 protein does not directly inhibit the caspase CED-3, in vitro
(2004) 11, 1309 1316 & 2004 Nature Publishing Group All rights reserved 1350-9047/04 $30.00 www.nature.com/cdd The Caenorhabditis elegans CED-9 protein does not directly inhibit the caspase CED-3, in vitro
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process
 RESEARCH ARTICLE 3305 Development 133, 3305-3315 (2006) doi:10.1242/dev.02495 The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic
RESEARCH ARTICLE 3305 Development 133, 3305-3315 (2006) doi:10.1242/dev.02495 The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic
REVIEW SESSION. Wednesday, September 15 5:30 PM SHANTZ 242 E
 REVIEW SESSION Wednesday, September 15 5:30 PM SHANTZ 242 E Gene Regulation Gene Regulation Gene expression can be turned on, turned off, turned up or turned down! For example, as test time approaches,
REVIEW SESSION Wednesday, September 15 5:30 PM SHANTZ 242 E Gene Regulation Gene Regulation Gene expression can be turned on, turned off, turned up or turned down! For example, as test time approaches,
Richik N. Ghosh, Linnette Grove, and Oleg Lapets ASSAY and Drug Development Technologies 2004, 2:
 1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
Exam 1 ID#: October 4, 2007
 Biology 4361 Name: KEY Exam 1 ID#: October 4, 2007 Multiple choice (one point each) (1-25) 1. The process of cells forming tissues and organs is called a. morphogenesis. b. differentiation. c. allometry.
Biology 4361 Name: KEY Exam 1 ID#: October 4, 2007 Multiple choice (one point each) (1-25) 1. The process of cells forming tissues and organs is called a. morphogenesis. b. differentiation. c. allometry.
Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290
 Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290 Question (from Introduction): How does svb control the
Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290 Question (from Introduction): How does svb control the
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Measuring TF-DNA interactions
 Measuring TF-DNA interactions How is Biological Complexity Achieved? Mediated by Transcription Factors (TFs) 2 Regulation of Gene Expression by Transcription Factors TF trans-acting factors TF TF TF TF
Measuring TF-DNA interactions How is Biological Complexity Achieved? Mediated by Transcription Factors (TFs) 2 Regulation of Gene Expression by Transcription Factors TF trans-acting factors TF TF TF TF
Control of Gene Expression
 Control of Gene Expression Mechanisms of Gene Control Gene Control in Eukaryotes Master Genes Gene Control In Prokaryotes Epigenetics Gene Expression The overall process by which information flows from
Control of Gene Expression Mechanisms of Gene Control Gene Control in Eukaryotes Master Genes Gene Control In Prokaryotes Epigenetics Gene Expression The overall process by which information flows from
T H E J O U R N A L O F C E L L B I O L O G Y
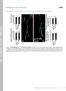 Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
