Estimation of Identification Methods of Gene Clusters Using GO Term Annotations from a Hierarchical Cluster Tree
|
|
|
- Randell Mathews
- 5 years ago
- Views:
Transcription
1 Estimation of Identification Methods of Gene Clusters Using GO Term Annotations from a Hierarchical Cluster Tree YOICHI YAMADA, YUKI MIYATA, MASANORI HIGASHIHARA*, KENJI SATOU Graduate School of Natural Science and Technology Kanazawa University Kakuma-machi, Kanazawa JAPAN * Graduate School of Knowledge Science Japan Advanced Institute of Science and Technology 1-1 Asahidai, Nomi, Ishikawa JAPAN Abstract: - The hierarchical clustering algorithm has frequently been applied to grouping genes sharing a certain characteristic from a microarray data set. Identification of clusters from a hierarchical cluster tree is generally conducted by cutting the tree at a certain level. In this method, the most parent clusters are identified as mutually correlated gene groups and their child clusters are ignored. However the child clusters have a possibility to show more significant GO term annotation than their parent clusters. To overcome this problem, Toronen developed a novel algorithm based on the calculation of each GO annotation in all the clusters that satisfy a threshold of correlation coefficient. However comparison of the algorithm with the general method have not been done enough so far. Therefore we compared the general method with Toronen s proposed algorithm for identifying overrepresented GO terms, and confirmed availability of the proposed algorithm. Key-Words: - Hierarchical clustering, Gene Ontology, Microarray data, Yeast cell cycle 1 Introduction Microarray technology allows us to monitor the expression of thousands of genes simultaneously [1]. As a first step for analyzing microarray data, a clustering algorithm is often applied and results in gene clusters sharing a certain characteristic [2]-[6]. For instance, time-course microarray experiments result in microarray data from a series of time points, and then the hierarchical clustering algorithm (HCA) is applied to the microarray data and gene groups showing a similar expression pattern across the time points are identified. Since genes grouped by the clustering algorism show similar expression patterns across a series of time points, they have a possibility to perform functionally related tasks: they may be functionally correlated. However, even if we know only the name of genes within the group, we will not understand what functional characteristics they share. In such cases, Gene Ontology (GO) is frequently used to give the genes any biological annotations [7]-[9]. The biological annotations are called GO terms and curated by GO Consortium. The GO terms annotate a number of genes from organisms of more than 50 species. Accordingly, GO terms commonly associated with genes in the same clusters are biological characteristics which they share. The significance of GO term annotation to genes is statistically tested [10]-[12]. Grouping of genes using microarray data is often performed by the HCA [2]. The identification of gene clusters by the HCA is generally performed by a break of merging of child clusters according to a threshold of correlation coefficient between genes. However disregard of child clusters of the identified clusters prevents us from detecting gene clusters with important characteristics. For instance, although statistical significance of GO term annotation to genes is influenced by the number of genes within a cluster, ISBN: ISSN
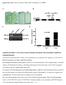
2 parent clusters always contain larger number of genes than their child clusters. Accordingly, child clusters have a possibility to show statistically significant GO annotations which their parent clusters do not show. In this context, Toronen reported a method in which statistical significance of each GO term annotation is examined in all the clusters (i.e., including both parent and child clusters) which satisfy a threshold of correlation coefficient between genes [13],[14]. However there are few reports including his report that show its availability based on comparison with the general method. In this study, we therefore compared the results from both methods using a yeast cell cycle microarray data set. 2 Problem Formulation Identification of clusters from a hierarchical cluster tree is generally performed by cutting the tree at a certain level. Fig. 1 shows an example of the process. The tree in Fig. 1 is cut at the level of the dotted line, and cluster 5, 8 and 9 are identified as clusters containing mutually correlated genes. In this case, other child clusters (1, 2, 3, 4, 6, 7) are ignored for the next analysis. In the next analysis, enrichment of each GO annotation to genes within the clusters is statistically estimated, and GO terms showing a significant p-value (e.g., p < 0.05) are assigned to the clusters. Here it is possible that a GO term show more statistically significant annotation in child clusters than in their parent clusters. For instance, although p-values of GO term a annotation in all the clusters below the dotted line are shown in Fig. 1, cluster 2, 6 and 7 show lower p-values than their parent clusters and also satisfy the criterion of under In this case, cluster 2, 6 and 7 should be identified for GO term a than cluster 8 and 9. In this context, Toronen reported a method in which statistical testing of each GO term annotation is conducted for all the clusters at lower level than a threshold of correlation coefficient, and the cluster showing the most significant p-value in the same branch of the cluster tree is selected for the GO term. It seems to be a reasonable method but there are few studies comparing the general method with Toronen s method. To examine availability of the method proposed by Toronen, we compared the general method with the proposed method using a yeast cell cycle microarray data set. Fig. 1: A hierarchical cluster tree of genes constructed from a microarray data set 3 Materials and methods 3.1 Microarray data set The microarray data set used in this study was produced by Spellman et al [15]. In the data set, yeast cells were alpha factor-arrested and synchronized, and periodically recovered in a series of time points after release. Then RNAs were extracted from recovered yeast cells. Control cells were also recovered from asynchronous yeast cells growing in the same culture condition at the same time points and their RNAs were extracted in the same way. Fluorescently labeled cdna was synthesized from each extracted RNA and the ratio of experimental to control cdna was measured every recovery time point. The expression ratio of each gene in obtained data was subjected to logarithmic conversion. 3.2 Construction of a hierarchical cluster tree from a microarray data set Cosine coefficient distances were calculated in all the possible gene pairs from the yeast cell cycle microarray data set. Centroid linkage method was applied to the formation of a hierarchical cluster tree using the open source clustering software, Cluster3 [16]. 3.3 Identification of clusters and their relevant GO terms from a hierarchical cluster tree by the general method ISBN: ISSN
3 A hierarchical cluster tree was cut at the level of each threshold (0.7, 0.75, 0.8, 0,85, 0.9) of correlation coefficient. Then the most parent clusters at the lower part of separated trees were identified. Next enrichment of GO term annotations to the identified clusters were calculated according to the following equation: min( n, M ) M C j N M Cn j p value = (1) C j N n where N is the number of genes examined by the microarray experiment which we refer to as population gene set, M is the number of genes annotated to a GO term in the population gene set, n is the number of genes within a cluster, and j is the number of genes assigned to the GO term in the cluster. GO term annotations of about 15,000 are calculated in each identified cluster. GO terms showing p-values under a threshold (e.g., 0.05) are identified as common characteristics among genes in each cluster. Although results of multiple comparisons require to be corrected, no correction is applied to those results because Bonferroni correction is so strict that a few GO terms show a statistical significance. 3.4 Identification of clusters and their relevant GO terms from a hierarchical cluster tree by Toronen s method A hierarchical cluster tree was cut at the level of each threshold (0.7, 0.75, 0.8, 0,85, 0.9) of correlation coefficient. The statistical enrichment of each GO annotation in all the clusters that fulfill each threshold (0.7, 0.75, 0.8, 0,85, 0.9) are calculated using the equation 1. GO term annotations of about 15,000 are calculated in each cluster. GO terms showing p-value below 0.05 are assigned to the clusters. Here repeated hits in the same branch of the cluster tree are discarded and the most statistically significant GO term is assigned to each branch of the cluster tree. Note that discarded GO terms can still be assigned to other parts of the cluster tree. 4 Results To compare results from general method and Toronen s method, gene clusters showing wide varieties of correlation coefficients were used. Eight clusters in Table 1 were identified by Spellman et al. using a Fourier algorithm and a correlation algorithm [15]. Genes within the clusters show periodically oscillated expression during the yeast cell cycle (see Fig. 2). Genes in each cluster show wide varieties of correlation coefficients in their expression patterns during the cell cycle (see Correlation coefficient in Table 1). For instance, although genes in Histone cluster show uniform expression patterns during the cell cycle, genes in MAT cluster show inconsistent expression patterns (see Fig. 2). Accordingly, comparison between the general method and Toronen s proposed method can be carried out using these clusters. For instance, if Toronen s proposed method has an ability to identify more overrepresented GO terms than general method, it will identify more GO terms significantly assigned to clusters with a high correlation coefficient than general method but not with a low correlation coefficient. To examine this supposition, we first determined significantly overrepresented GO terms (p < 0.05) in the clusters identified by Spellman et al., as shown in Table 1. A B Fig. 2: Expression of genes in (A) Histone cluster and (B) MAT cluster at each time point. ISBN: ISSN
4 Table 1: Comparison of the general method and Toronen s proposed method Correlation Overrepresented Threshold of correlation coefficient Gene cluster coefficient GO term CLN2 cluster /44 61/40 61/61 62/44 61/45 Y` cluster /0 1/0 1/0 1/1 1/1 MAT cluster /0 0/0 0/0 0/0 9/9 MCM cluster /17 30/17 21/16 6/4 2/2 SIC1 cluster /7 15/8 17/9 17/14 14/13 CLB2 cluster /0 28/0 34/25 34/25 32/21 Histone cluster /30 32/27 32/27 32/27 32/30 MET cluster /14 19/18 7/7 0/0 0/0 In Table 1, Overrepresented GO term shows the number of GO terms significantly associated with each Gene cluster. Numbers to the left and the right of each slash in Table 1 represent the numbers of the overrepresented GO term identified by Toronen s method and the general method, respectively. Note that not only the overrepresented GO terms but also genes annotated to those overlapped between clusters identified by Spellman et al. and those identified by hierarchical clustering algorithm. In most of thresholds of correlation coefficient, Toronen s method could identify more overrepresented GO terms in all the gene clusters showing positive correlation coefficients than general method (see Table 1). In contrast, MAT cluster showing the negative correlation coefficient displayed no difference between Toronen s method and the general method in all thresholds of correlation coefficient (see Table 1). 5 Conclusion In this study, we compared two methods (i.e., the general method and Toronen s method) for identification of clusters and their relevant GO terms from a hierarchical cluster tree. As a result, Toronen s method could identify more GO terms significantly associated with gene clusters showing positive correlation coefficients than the general method. Moreover, the number of GO terms significantly associated with gene clusters showing the negative correlation coefficient is identical between Toronen s method and the general method: identification rate of false positive is identical between both methods. Consequently, our simulation confirmed availability of Toronen s method for identification of clusters and their relevant GO terms from a hierarchical cluster tree. References: [1] D. Shalon, S.J. Smith, P.O. Brown, A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization, Genome Res., Vol.6, 1996, pp [2] M.B. Eisen, P.T. Spellman, P.O. Brown, D. Botstein, Cluster analysis and display of genome-wide expression patterns, Proc. Natl. Acad. Sci. USA, Vol.95, 1998, pp [3] P. Tamayo, D. Slonim, J. Mesirov, Q. Zhu, S. Kitareewan, E. Dmitrovsky, E.S. Lander, T.R. Golub, Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation, Proc. Natl. Acad. Sci. USA,. Vol.96, 1999, pp [4] K.Y. Yeung, C. Fraley, A. Murua, A.E. Raftery, W.L. Ruzzo, Model-based clustering and data transformations for gene expression data, Bioinformatics, Vol.17, 2001, pp [5] S. Datta and S. Datta, Methods for evaluating clustering algorithms for gene expression data using a reference set of functional classes, BMC Bioinformatics, Vol.7, 2006, pp.397. [6] F.D. Gibbons, F.P. Roth, Judging the quality of gene expression-based clustering methods using gene annotation, Genome Res., Vol. 12, 2002, pp [7] M. Ashburner, C.A. Ball, J.A. Blake, D. Botstein, H. Butler, J.M. Cherry, A.P. Davis, K. Dolinski, S.S. Dwight, J.T. Eppig, M.A. Harris, D.P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J.C. Matese, ISBN: ISSN
5 J.E. Richardson, M. Ringwald, G.M. Rubin, G. Sherlock, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nat. Genet., Vol.25, 2000, pp [8] Gene Ontology Consortium, Creating the gene ontology resource: design and implementation, Genome Res., Vo.11, 2001, pp [9] E. Camon, M. Magrane, D. Barrell, V. Lee, E. Dimmer, J. Maslen, D. Binns, N, Harte, R. Lopez and R. Apweiler, The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology, Nucleic Acids Res., Vol.32 (Database issue), 2004b, D [10] T. Beissbarth, T. P. Speed, GOstat: find statistically overrepresented Gene Ontologies within a group of genes, Bioinformatics, Vol.20, 2004, pp [11] E. I. Boyle, S. Weng, J. Gollub, H. Jin, D. Botstein, J. M. Cherry, G. Sherlock, GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes, Bioinformatics, Vol.20, 2004, pp [12] R.S. Sealfon, M.A. Hibbs, C. Huttenhower, C.L. Myers, O.G. Troyanskaya, GOLEM: an interactive graph-based gene-ontology navigation and analysis tool, BMC Bioinformatics, Vol.7, 2006, pp.443. [13] P. Toronen, Selection of informative clusters from hierarchical cluster tree with gene classes, BMC Bioinformatics, Vol.5, 2004, pp.32. [14] M. Kankainen, G. Brader, P. Törönen, E.T. Palva, L. Holm, Identifying functional gene sets from hierarchically clustered expression data: map of abiotic stress regulated genes in Arabidopsis thaliana, Nucleic Acids Res., Vol.34, 2006, e124. [15] P. T. Spellman, G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M.B. Eisen, P.O. Brown, D. Botstein, B. Futcher, Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization, Mol. Biol. Cell, Vol.9, 1998 pp [16] M.J.L. de Hoon, S. Imoto, J. Nolan, S. Miyano, Open source clustering software, Bioinformatics, Vol.20, 2004, pp ISBN: ISSN
Analyzing Microarray Time course Genome wide Data
 OR 779 Functional Data Analysis Course Project Analyzing Microarray Time course Genome wide Data Presented by Xin Zhao April 29, 2002 Cornell University Overview 1. Introduction Biological Background Biological
OR 779 Functional Data Analysis Course Project Analyzing Microarray Time course Genome wide Data Presented by Xin Zhao April 29, 2002 Cornell University Overview 1. Introduction Biological Background Biological
Cluster Analysis of Gene Expression Microarray Data. BIOL 495S/ CS 490B/ MATH 490B/ STAT 490B Introduction to Bioinformatics April 8, 2002
 Cluster Analysis of Gene Expression Microarray Data BIOL 495S/ CS 490B/ MATH 490B/ STAT 490B Introduction to Bioinformatics April 8, 2002 1 Data representations Data are relative measurements log 2 ( red
Cluster Analysis of Gene Expression Microarray Data BIOL 495S/ CS 490B/ MATH 490B/ STAT 490B Introduction to Bioinformatics April 8, 2002 1 Data representations Data are relative measurements log 2 ( red
Inferring Gene Regulatory Networks from Time-Ordered Gene Expression Data of Bacillus Subtilis Using Differential Equations
 Inferring Gene Regulatory Networks from Time-Ordered Gene Expression Data of Bacillus Subtilis Using Differential Equations M.J.L. de Hoon, S. Imoto, K. Kobayashi, N. Ogasawara, S. Miyano Pacific Symposium
Inferring Gene Regulatory Networks from Time-Ordered Gene Expression Data of Bacillus Subtilis Using Differential Equations M.J.L. de Hoon, S. Imoto, K. Kobayashi, N. Ogasawara, S. Miyano Pacific Symposium
 Supplemental Table 1. Primers used for qrt-pcr analysis Name abf2rtf abf2rtr abf3rtf abf3rtf abf4rtf abf4rtr ANAC096RTF ANAC096RTR abf2lp abf2rp abf3lp abf3lp abf4lp abf4rp ANAC096F(XbaI) ANAC096R(BamHI)
Supplemental Table 1. Primers used for qrt-pcr analysis Name abf2rtf abf2rtr abf3rtf abf3rtf abf4rtf abf4rtr ANAC096RTF ANAC096RTR abf2lp abf2rp abf3lp abf3lp abf4lp abf4rp ANAC096F(XbaI) ANAC096R(BamHI)
GOSAP: Gene Ontology Based Semantic Alignment of Biological Pathways
 GOSAP: Gene Ontology Based Semantic Alignment of Biological Pathways Jonas Gamalielsson and Björn Olsson Systems Biology Group, Skövde University, Box 407, Skövde, 54128, Sweden, [jonas.gamalielsson][bjorn.olsson]@his.se,
GOSAP: Gene Ontology Based Semantic Alignment of Biological Pathways Jonas Gamalielsson and Björn Olsson Systems Biology Group, Skövde University, Box 407, Skövde, 54128, Sweden, [jonas.gamalielsson][bjorn.olsson]@his.se,
Supplemental Information for Pramila et al. Periodic Normal Mixture Model (PNM)
 Supplemental Information for Pramila et al. Periodic Normal Mixture Model (PNM) The data sets alpha30 and alpha38 were analyzed with PNM (Lu et al. 2004). The first two time points were deleted to alleviate
Supplemental Information for Pramila et al. Periodic Normal Mixture Model (PNM) The data sets alpha30 and alpha38 were analyzed with PNM (Lu et al. 2004). The first two time points were deleted to alleviate
Bioinformatics. Transcriptome
 Bioinformatics Transcriptome Jacques.van.Helden@ulb.ac.be Université Libre de Bruxelles, Belgique Laboratoire de Bioinformatique des Génomes et des Réseaux (BiGRe) http://www.bigre.ulb.ac.be/ Bioinformatics
Bioinformatics Transcriptome Jacques.van.Helden@ulb.ac.be Université Libre de Bruxelles, Belgique Laboratoire de Bioinformatique des Génomes et des Réseaux (BiGRe) http://www.bigre.ulb.ac.be/ Bioinformatics
Package neat. February 23, 2018
 Type Package Title Efficient Network Enrichment Analysis Test Version 1.1.3 Date 2018-02-23 Depends R (>= 3.3.0) Package neat February 23, 2018 Author Mirko Signorelli, Veronica Vinciotti and Ernst C.
Type Package Title Efficient Network Enrichment Analysis Test Version 1.1.3 Date 2018-02-23 Depends R (>= 3.3.0) Package neat February 23, 2018 Author Mirko Signorelli, Veronica Vinciotti and Ernst C.
Advances in microarray technologies (1 5) have enabled
 Statistical modeling of large microarray data sets to identify stimulus-response profiles Lue Ping Zhao*, Ross Prentice*, and Linda Breeden Divisions of *Public Health Sciences and Basic Sciences, Fred
Statistical modeling of large microarray data sets to identify stimulus-response profiles Lue Ping Zhao*, Ross Prentice*, and Linda Breeden Divisions of *Public Health Sciences and Basic Sciences, Fred
Modelling Gene Expression Data over Time: Curve Clustering with Informative Prior Distributions.
 BAYESIAN STATISTICS 7, pp. 000 000 J. M. Bernardo, M. J. Bayarri, J. O. Berger, A. P. Dawid, D. Heckerman, A. F. M. Smith and M. West (Eds.) c Oxford University Press, 2003 Modelling Data over Time: Curve
BAYESIAN STATISTICS 7, pp. 000 000 J. M. Bernardo, M. J. Bayarri, J. O. Berger, A. P. Dawid, D. Heckerman, A. F. M. Smith and M. West (Eds.) c Oxford University Press, 2003 Modelling Data over Time: Curve
Exploratory statistical analysis of multi-species time course gene expression
 Exploratory statistical analysis of multi-species time course gene expression data Eng, Kevin H. University of Wisconsin, Department of Statistics 1300 University Avenue, Madison, WI 53706, USA. E-mail:
Exploratory statistical analysis of multi-species time course gene expression data Eng, Kevin H. University of Wisconsin, Department of Statistics 1300 University Avenue, Madison, WI 53706, USA. E-mail:
Classification of Gene Expression Profiles: Comparison of K-means and Expectation Maximization Algorithms
 Eighth International Conference on Hybrid Intelligent Systems Classification of Gene Expression Profiles: Comparison of K-means and Expectation Maximization Algorithms Cristina Rubio-Escudero Dpto. Lenguajes
Eighth International Conference on Hybrid Intelligent Systems Classification of Gene Expression Profiles: Comparison of K-means and Expectation Maximization Algorithms Cristina Rubio-Escudero Dpto. Lenguajes
IMPORTANCE OF SECONDARY STRUCTURE ELEMENTS FOR PREDICTION OF GO ANNOTATIONS
 IMPORTANCE OF SECONDARY STRUCTURE ELEMENTS FOR PREDICTION OF GO ANNOTATIONS Aslı Filiz 1, Eser Aygün 2, Özlem Keskin 3 and Zehra Cataltepe 2 1 Informatics Institute and 2 Computer Engineering Department,
IMPORTANCE OF SECONDARY STRUCTURE ELEMENTS FOR PREDICTION OF GO ANNOTATIONS Aslı Filiz 1, Eser Aygün 2, Özlem Keskin 3 and Zehra Cataltepe 2 1 Informatics Institute and 2 Computer Engineering Department,
Lecture 5: November 19, Minimizing the maximum intracluster distance
 Analysis of DNA Chips and Gene Networks Spring Semester, 2009 Lecture 5: November 19, 2009 Lecturer: Ron Shamir Scribe: Renana Meller 5.1 Minimizing the maximum intracluster distance 5.1.1 Introduction
Analysis of DNA Chips and Gene Networks Spring Semester, 2009 Lecture 5: November 19, 2009 Lecturer: Ron Shamir Scribe: Renana Meller 5.1 Minimizing the maximum intracluster distance 5.1.1 Introduction
ProtoNet 4.0: A hierarchical classification of one million protein sequences
 ProtoNet 4.0: A hierarchical classification of one million protein sequences Noam Kaplan 1*, Ori Sasson 2, Uri Inbar 2, Moriah Friedlich 2, Menachem Fromer 2, Hillel Fleischer 2, Elon Portugaly 2, Nathan
ProtoNet 4.0: A hierarchical classification of one million protein sequences Noam Kaplan 1*, Ori Sasson 2, Uri Inbar 2, Moriah Friedlich 2, Menachem Fromer 2, Hillel Fleischer 2, Elon Portugaly 2, Nathan
CLUSTER, FUNCTION AND PROMOTER: ANALYSIS OF YEAST EXPRESSION ARRAY
 CLUSTER, FUNCTION AND PROMOTER: ANALYSIS OF YEAST EXPRESSION ARRAY J. ZHU, M. Q. ZHANG Cold Spring Harbor Lab, P. O. Box 100 Cold Spring Harbor, NY 11724 Gene clusters could be derived based on expression
CLUSTER, FUNCTION AND PROMOTER: ANALYSIS OF YEAST EXPRESSION ARRAY J. ZHU, M. Q. ZHANG Cold Spring Harbor Lab, P. O. Box 100 Cold Spring Harbor, NY 11724 Gene clusters could be derived based on expression
Genome-wide Analysis of Functions Regulated by Sets of Transcription Factors
 Genome-wide Analysis of Functions Regulated by Sets of Transcription Factors Szymon M. Kiełbasa 1, Nils Blüthgen, Hanspeter Herzel Humboldt University, Institute for Theoretical Biology, Invalidenstraße
Genome-wide Analysis of Functions Regulated by Sets of Transcription Factors Szymon M. Kiełbasa 1, Nils Blüthgen, Hanspeter Herzel Humboldt University, Institute for Theoretical Biology, Invalidenstraße
Topographic Independent Component Analysis of Gene Expression Time Series Data
 Topographic Independent Component Analysis of Gene Expression Time Series Data Sookjeong Kim and Seungjin Choi Department of Computer Science Pohang University of Science and Technology San 31 Hyoja-dong,
Topographic Independent Component Analysis of Gene Expression Time Series Data Sookjeong Kim and Seungjin Choi Department of Computer Science Pohang University of Science and Technology San 31 Hyoja-dong,
Integration of functional genomics data
 Integration of functional genomics data Laboratoire Bordelais de Recherche en Informatique (UMR) Centre de Bioinformatique de Bordeaux (Plateforme) Rennes Oct. 2006 1 Observations and motivations Genomics
Integration of functional genomics data Laboratoire Bordelais de Recherche en Informatique (UMR) Centre de Bioinformatique de Bordeaux (Plateforme) Rennes Oct. 2006 1 Observations and motivations Genomics
Constructing Signal Transduction Networks Using Multiple Signaling Feature Data
 Constructing Signal Transduction Networks Using Multiple Signaling Feature Data Thanh-Phuong Nguyen 1, Kenji Satou 2, Tu-Bao Ho 1 and Katsuhiko Takabayashi 3 1 Japan Advanced Institute of Science and Technology
Constructing Signal Transduction Networks Using Multiple Signaling Feature Data Thanh-Phuong Nguyen 1, Kenji Satou 2, Tu-Bao Ho 1 and Katsuhiko Takabayashi 3 1 Japan Advanced Institute of Science and Technology
Modeling Gene Expression from Microarray Expression Data with State-Space Equations. F.X. Wu, W.J. Zhang, and A.J. Kusalik
 Modeling Gene Expression from Microarray Expression Data with State-Space Equations FX Wu, WJ Zhang, and AJ Kusalik Pacific Symposium on Biocomputing 9:581-592(2004) MODELING GENE EXPRESSION FROM MICROARRAY
Modeling Gene Expression from Microarray Expression Data with State-Space Equations FX Wu, WJ Zhang, and AJ Kusalik Pacific Symposium on Biocomputing 9:581-592(2004) MODELING GENE EXPRESSION FROM MICROARRAY
Regression Model In The Analysis Of Micro Array Data-Gene Expression Detection
 Jamal Fathima.J.I 1 and P.Venkatesan 1. Research Scholar -Department of statistics National Institute For Research In Tuberculosis, Indian Council For Medical Research,Chennai,India,.Department of statistics
Jamal Fathima.J.I 1 and P.Venkatesan 1. Research Scholar -Department of statistics National Institute For Research In Tuberculosis, Indian Council For Medical Research,Chennai,India,.Department of statistics
Genome-Scale Gene Function Prediction Using Multiple Sources of High-Throughput Data in Yeast Saccharomyces cerevisiae ABSTRACT
 OMICS A Journal of Integrative Biology Volume 8, Number 4, 2004 Mary Ann Liebert, Inc. Genome-Scale Gene Function Prediction Using Multiple Sources of High-Throughput Data in Yeast Saccharomyces cerevisiae
OMICS A Journal of Integrative Biology Volume 8, Number 4, 2004 Mary Ann Liebert, Inc. Genome-Scale Gene Function Prediction Using Multiple Sources of High-Throughput Data in Yeast Saccharomyces cerevisiae
Towards Detecting Protein Complexes from Protein Interaction Data
 Towards Detecting Protein Complexes from Protein Interaction Data Pengjun Pei 1 and Aidong Zhang 1 Department of Computer Science and Engineering State University of New York at Buffalo Buffalo NY 14260,
Towards Detecting Protein Complexes from Protein Interaction Data Pengjun Pei 1 and Aidong Zhang 1 Department of Computer Science and Engineering State University of New York at Buffalo Buffalo NY 14260,
PROTEIN FUNCTION PREDICTION WITH AMINO ACID SEQUENCE AND SECONDARY STRUCTURE ALIGNMENT SCORES
 PROTEIN FUNCTION PREDICTION WITH AMINO ACID SEQUENCE AND SECONDARY STRUCTURE ALIGNMENT SCORES Eser Aygün 1, Caner Kömürlü 2, Zafer Aydin 3 and Zehra Çataltepe 1 1 Computer Engineering Department and 2
PROTEIN FUNCTION PREDICTION WITH AMINO ACID SEQUENCE AND SECONDARY STRUCTURE ALIGNMENT SCORES Eser Aygün 1, Caner Kömürlü 2, Zafer Aydin 3 and Zehra Çataltepe 1 1 Computer Engineering Department and 2
Mixture models for analysing transcriptome and ChIP-chip data
 Mixture models for analysing transcriptome and ChIP-chip data Marie-Laure Martin-Magniette French National Institute for agricultural research (INRA) Unit of Applied Mathematics and Informatics at AgroParisTech,
Mixture models for analysing transcriptome and ChIP-chip data Marie-Laure Martin-Magniette French National Institute for agricultural research (INRA) Unit of Applied Mathematics and Informatics at AgroParisTech,
Tree-Dependent Components of Gene Expression Data for Clustering
 Tree-Dependent Components of Gene Expression Data for Clustering Jong Kyoung Kim and Seungjin Choi Department of Computer Science Pohang University of Science and Technology San 31 Hyoja-dong, Nam-gu Pohang
Tree-Dependent Components of Gene Expression Data for Clustering Jong Kyoung Kim and Seungjin Choi Department of Computer Science Pohang University of Science and Technology San 31 Hyoja-dong, Nam-gu Pohang
Sparse regularization for functional logistic regression models
 Sparse regularization for functional logistic regression models Hidetoshi Matsui The Center for Data Science Education and Research, Shiga University 1-1-1 Banba, Hikone, Shiga, 5-85, Japan. hmatsui@biwako.shiga-u.ac.jp
Sparse regularization for functional logistic regression models Hidetoshi Matsui The Center for Data Science Education and Research, Shiga University 1-1-1 Banba, Hikone, Shiga, 5-85, Japan. hmatsui@biwako.shiga-u.ac.jp
A New Method to Build Gene Regulation Network Based on Fuzzy Hierarchical Clustering Methods
 International Academic Institute for Science and Technology International Academic Journal of Science and Engineering Vol. 3, No. 6, 2016, pp. 169-176. ISSN 2454-3896 International Academic Journal of
International Academic Institute for Science and Technology International Academic Journal of Science and Engineering Vol. 3, No. 6, 2016, pp. 169-176. ISSN 2454-3896 International Academic Journal of
Fuzzy Clustering of Gene Expression Data
 Fuzzy Clustering of Gene Data Matthias E. Futschik and Nikola K. Kasabov Department of Information Science, University of Otago P.O. Box 56, Dunedin, New Zealand email: mfutschik@infoscience.otago.ac.nz,
Fuzzy Clustering of Gene Data Matthias E. Futschik and Nikola K. Kasabov Department of Information Science, University of Otago P.O. Box 56, Dunedin, New Zealand email: mfutschik@infoscience.otago.ac.nz,
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Application of random matrix theory to microarray data for discovering functional gene modules
 Application of random matrix theory to microarray data for discovering functional gene modules Feng Luo, 1 Jianxin Zhong, 2,3, * Yunfeng Yang, 4 and Jizhong Zhou 4,5, 1 Department of Computer Science,
Application of random matrix theory to microarray data for discovering functional gene modules Feng Luo, 1 Jianxin Zhong, 2,3, * Yunfeng Yang, 4 and Jizhong Zhou 4,5, 1 Department of Computer Science,
Francisco M. Couto Mário J. Silva Pedro Coutinho
 Francisco M. Couto Mário J. Silva Pedro Coutinho DI FCUL TR 03 29 Departamento de Informática Faculdade de Ciências da Universidade de Lisboa Campo Grande, 1749 016 Lisboa Portugal Technical reports are
Francisco M. Couto Mário J. Silva Pedro Coutinho DI FCUL TR 03 29 Departamento de Informática Faculdade de Ciências da Universidade de Lisboa Campo Grande, 1749 016 Lisboa Portugal Technical reports are
The Continuum Model of the Eukaryotic Cell Cycle: Application to G1-phase control, Rb phosphorylation, Microarray Analysis of Gene Expression,
 The Continuum Model of the Eukaryotic Cell Cycle: Application to G1-phase control, Rb phosphorylation, Microarray Analysis of Gene Expression, and Cell Synchronization Stephen Cooper Department of Microbiology
The Continuum Model of the Eukaryotic Cell Cycle: Application to G1-phase control, Rb phosphorylation, Microarray Analysis of Gene Expression, and Cell Synchronization Stephen Cooper Department of Microbiology
2 GENE FUNCTIONAL SIMILARITY. 2.1 Semantic values of GO terms
 Bioinformatics Advance Access published March 7, 2007 The Author (2007). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Bioinformatics Advance Access published March 7, 2007 The Author (2007). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Keywords: microarray analysis, data integration, function prediction, biological networks, pathway prediction
 Olga Troyanskaya is an Assistant Professor at the Department of Computer Science and in the Lewis-Sigler Institute for Integrative Genomics at Princeton University. Her laboratory focuses on prediction
Olga Troyanskaya is an Assistant Professor at the Department of Computer Science and in the Lewis-Sigler Institute for Integrative Genomics at Princeton University. Her laboratory focuses on prediction
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Breker et al., http://www.jcb.org/cgi/content/full/jcb.201301120/dc1 Figure S1. Single-cell proteomics of stress responses. (a) Using
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Breker et al., http://www.jcb.org/cgi/content/full/jcb.201301120/dc1 Figure S1. Single-cell proteomics of stress responses. (a) Using
Dynamics of cellular level function and regulation derived from murine expression array data
 Dynamics of cellular level function and regulation derived from murine expression array data Benjamin de Bivort*, Sui Huang, and Yaneer Bar-Yam* *Department of Molecular and Cellular Biology, Harvard University,
Dynamics of cellular level function and regulation derived from murine expression array data Benjamin de Bivort*, Sui Huang, and Yaneer Bar-Yam* *Department of Molecular and Cellular Biology, Harvard University,
BMD645. Integration of Omics
 BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
Computational methods for predicting protein-protein interactions
 Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
The Generalized Topological Overlap Matrix For Detecting Modules in Gene Networks
 The Generalized Topological Overlap Matrix For Detecting Modules in Gene Networks Andy M. Yip Steve Horvath Abstract Systems biologic studies of gene and protein interaction networks have found that these
The Generalized Topological Overlap Matrix For Detecting Modules in Gene Networks Andy M. Yip Steve Horvath Abstract Systems biologic studies of gene and protein interaction networks have found that these
Data visualization and clustering: an application to gene expression data
 Data visualization and clustering: an application to gene expression data Francesco Napolitano Università degli Studi di Salerno Dipartimento di Matematica e Informatica DAA Erice, April 2007 Thanks to
Data visualization and clustering: an application to gene expression data Francesco Napolitano Università degli Studi di Salerno Dipartimento di Matematica e Informatica DAA Erice, April 2007 Thanks to
Shrinkage-Based Similarity Metric for Cluster Analysis of Microarray Data
 Shrinkage-Based Similarity Metric for Cluster Analysis of Microarray Data Vera Cherepinsky, Jiawu Feng, Marc Rejali, and Bud Mishra, Courant Institute, New York University, 5 Mercer Street, New York, NY
Shrinkage-Based Similarity Metric for Cluster Analysis of Microarray Data Vera Cherepinsky, Jiawu Feng, Marc Rejali, and Bud Mishra, Courant Institute, New York University, 5 Mercer Street, New York, NY
6.047 / Computational Biology: Genomes, Networks, Evolution Fall 2008
 MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu 6.047 / 6.878 Computational Biology: Genomes, Networks, Evolution Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Multiplicative background correction for spotted. microarrays to improve reproducibility
 Multiplicative background correction for spotted microarrays to improve reproducibility DABAO ZHANG,, MIN ZHANG, MARTIN T. WELLS, March 12, 2006 Department of Statistics, Purdue University, West Lafayette,
Multiplicative background correction for spotted microarrays to improve reproducibility DABAO ZHANG,, MIN ZHANG, MARTIN T. WELLS, March 12, 2006 Department of Statistics, Purdue University, West Lafayette,
Improving the identification of differentially expressed genes in cdna microarray experiments
 Improving the identification of differentially expressed genes in cdna microarray experiments Databionics Research Group University of Marburg 33 Marburg, Germany Alfred Ultsch Abstract. The identification
Improving the identification of differentially expressed genes in cdna microarray experiments Databionics Research Group University of Marburg 33 Marburg, Germany Alfred Ultsch Abstract. The identification
Learning Causal Networks from Microarray Data
 Learning Causal Networks from Microarray Data Nasir Ahsan Michael Bain John Potter Bruno Gaëta Mark Temple Ian Dawes School of Computer Science and Engineering School of Biotechnology and Biomolecular
Learning Causal Networks from Microarray Data Nasir Ahsan Michael Bain John Potter Bruno Gaëta Mark Temple Ian Dawes School of Computer Science and Engineering School of Biotechnology and Biomolecular
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Comparison of Human Protein-Protein Interaction Maps
 Comparison of Human Protein-Protein Interaction Maps Matthias E. Futschik 1, Gautam Chaurasia 1,2, Erich Wanker 2 and Hanspeter Herzel 1 1 Institute for Theoretical Biology, Charité, Humboldt-Universität
Comparison of Human Protein-Protein Interaction Maps Matthias E. Futschik 1, Gautam Chaurasia 1,2, Erich Wanker 2 and Hanspeter Herzel 1 1 Institute for Theoretical Biology, Charité, Humboldt-Universität
arxiv: v1 [q-bio.mn] 7 Nov 2018
![arxiv: v1 [q-bio.mn] 7 Nov 2018 arxiv: v1 [q-bio.mn] 7 Nov 2018](/thumbs/90/102608433.jpg) Role of self-loop in cell-cycle network of budding yeast Shu-ichi Kinoshita a, Hiroaki S. Yamada b a Department of Mathematical Engineering, Faculty of Engeneering, Musashino University, -- Ariake Koutou-ku,
Role of self-loop in cell-cycle network of budding yeast Shu-ichi Kinoshita a, Hiroaki S. Yamada b a Department of Mathematical Engineering, Faculty of Engeneering, Musashino University, -- Ariake Koutou-ku,
Genomics and bioinformatics summary. Finding genes -- computer searches
 Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
Comparison of Protein-Protein Interaction Confidence Assignment Schemes
 Comparison of Protein-Protein Interaction Confidence Assignment Schemes Silpa Suthram 1, Tomer Shlomi 2, Eytan Ruppin 2, Roded Sharan 2, and Trey Ideker 1 1 Department of Bioengineering, University of
Comparison of Protein-Protein Interaction Confidence Assignment Schemes Silpa Suthram 1, Tomer Shlomi 2, Eytan Ruppin 2, Roded Sharan 2, and Trey Ideker 1 1 Department of Bioengineering, University of
Bioinformatics 2. Yeast two hybrid. Proteomics. Proteomics
 GENOME Bioinformatics 2 Proteomics protein-gene PROTEOME protein-protein METABOLISM Slide from http://www.nd.edu/~networks/ Citrate Cycle Bio-chemical reactions What is it? Proteomics Reveal protein Protein
GENOME Bioinformatics 2 Proteomics protein-gene PROTEOME protein-protein METABOLISM Slide from http://www.nd.edu/~networks/ Citrate Cycle Bio-chemical reactions What is it? Proteomics Reveal protein Protein
A Case Study -- Chu et al. The Transcriptional Program of Sporulation in Budding Yeast. What is Sporulation? CSE 527, W.L. Ruzzo 1
 A Case Study -- Chu et al. An interesting early microarray paper My goals Show arrays used in a real experiment Show where computation is important Start looking at analysis techniques The Transcriptional
A Case Study -- Chu et al. An interesting early microarray paper My goals Show arrays used in a real experiment Show where computation is important Start looking at analysis techniques The Transcriptional
Robust Community Detection Methods with Resolution Parameter for Complex Detection in Protein Protein Interaction Networks
 Robust Community Detection Methods with Resolution Parameter for Complex Detection in Protein Protein Interaction Networks Twan van Laarhoven and Elena Marchiori Institute for Computing and Information
Robust Community Detection Methods with Resolution Parameter for Complex Detection in Protein Protein Interaction Networks Twan van Laarhoven and Elena Marchiori Institute for Computing and Information
Superstability of the yeast cell-cycle dynamics: Ensuring causality in the presence of biochemical stochasticity
 Journal of Theoretical Biology 245 (27) 638 643 www.elsevier.com/locate/yjtbi Superstability of the yeast cell-cycle dynamics: Ensuring causality in the presence of biochemical stochasticity Stefan Braunewell,
Journal of Theoretical Biology 245 (27) 638 643 www.elsevier.com/locate/yjtbi Superstability of the yeast cell-cycle dynamics: Ensuring causality in the presence of biochemical stochasticity Stefan Braunewell,
Gene Ontology and Functional Enrichment. Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein
 Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Missing Value Estimation for Time Series Microarray Data Using Linear Dynamical Systems Modeling
 22nd International Conference on Advanced Information Networking and Applications - Workshops Missing Value Estimation for Time Series Microarray Data Using Linear Dynamical Systems Modeling Connie Phong
22nd International Conference on Advanced Information Networking and Applications - Workshops Missing Value Estimation for Time Series Microarray Data Using Linear Dynamical Systems Modeling Connie Phong
Written Exam 15 December Course name: Introduction to Systems Biology Course no
 Technical University of Denmark Written Exam 15 December 2008 Course name: Introduction to Systems Biology Course no. 27041 Aids allowed: Open book exam Provide your answers and calculations on separate
Technical University of Denmark Written Exam 15 December 2008 Course name: Introduction to Systems Biology Course no. 27041 Aids allowed: Open book exam Provide your answers and calculations on separate
Procedure to Create NCBI KOGS
 Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
Clustering Analysis of SAGE Transcription Profiles Using a Poisson Approach
 Boo_Nielsen_886768_Proof1_February 6, Clustering Analysis of SAGE Transcription Profiles Using a Poisson Approach Haiyan Huang, Li Cai, and Wing H. Wong Summary To gain insights into the biological function
Boo_Nielsen_886768_Proof1_February 6, Clustering Analysis of SAGE Transcription Profiles Using a Poisson Approach Haiyan Huang, Li Cai, and Wing H. Wong Summary To gain insights into the biological function
Improving Gene Functional Analysis in Ethylene-induced Leaf Abscission using GO and ProteInOn
 Improving Gene Functional Analysis in Ethylene-induced Leaf Abscission using GO and ProteInOn Sara Domingos 1, Cátia Pesquita 2, Francisco M. Couto 2, Luis F. Goulao 3, Cristina Oliveira 1 1 Instituto
Improving Gene Functional Analysis in Ethylene-induced Leaf Abscission using GO and ProteInOn Sara Domingos 1, Cátia Pesquita 2, Francisco M. Couto 2, Luis F. Goulao 3, Cristina Oliveira 1 1 Instituto
Genome Annotation. Bioinformatics and Computational Biology. Genome sequencing Assembly. Gene prediction. Protein targeting.
 Genome Annotation Bioinformatics and Computational Biology Genome Annotation Frank Oliver Glöckner 1 Genome Analysis Roadmap Genome sequencing Assembly Gene prediction Protein targeting trna prediction
Genome Annotation Bioinformatics and Computational Biology Genome Annotation Frank Oliver Glöckner 1 Genome Analysis Roadmap Genome sequencing Assembly Gene prediction Protein targeting trna prediction
Small RNA in rice genome
 Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
Comparative Network Analysis
 Comparative Network Analysis BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2016 Anthony Gitter gitter@biostat.wisc.edu These slides, excluding third-party material, are licensed under CC BY-NC 4.0 by
Comparative Network Analysis BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2016 Anthony Gitter gitter@biostat.wisc.edu These slides, excluding third-party material, are licensed under CC BY-NC 4.0 by
Introduction to clustering methods for gene expression data analysis
 Introduction to clustering methods for gene expression data analysis Giorgio Valentini e-mail: valentini@dsi.unimi.it Outline Levels of analysis of DNA microarray data Clustering methods for functional
Introduction to clustering methods for gene expression data analysis Giorgio Valentini e-mail: valentini@dsi.unimi.it Outline Levels of analysis of DNA microarray data Clustering methods for functional
Rule learning for gene expression data
 Rule learning for gene expression data Stefan Enroth Original slides by Torgeir R. Hvidsten The Linnaeus Centre for Bioinformatics Predicting biological process from gene expression time profiles Papers:
Rule learning for gene expression data Stefan Enroth Original slides by Torgeir R. Hvidsten The Linnaeus Centre for Bioinformatics Predicting biological process from gene expression time profiles Papers:
INCORPORATING GRAPH FEATURES FOR PREDICTING PROTEIN-PROTEIN INTERACTIONS
 INCORPORATING GRAPH FEATURES FOR PREDICTING PROTEIN-PROTEIN INTERACTIONS Martin S. R. Paradesi Department of Computing and Information Sciences, Kansas State University 234 Nichols Hall, Manhattan, KS
INCORPORATING GRAPH FEATURES FOR PREDICTING PROTEIN-PROTEIN INTERACTIONS Martin S. R. Paradesi Department of Computing and Information Sciences, Kansas State University 234 Nichols Hall, Manhattan, KS
Learning in Bayesian Networks
 Learning in Bayesian Networks Florian Markowetz Max-Planck-Institute for Molecular Genetics Computational Molecular Biology Berlin Berlin: 20.06.2002 1 Overview 1. Bayesian Networks Stochastic Networks
Learning in Bayesian Networks Florian Markowetz Max-Planck-Institute for Molecular Genetics Computational Molecular Biology Berlin Berlin: 20.06.2002 1 Overview 1. Bayesian Networks Stochastic Networks
Discovering molecular pathways from protein interaction and ge
 Discovering molecular pathways from protein interaction and gene expression data 9-4-2008 Aim To have a mechanism for inferring pathways from gene expression and protein interaction data. Motivation Why
Discovering molecular pathways from protein interaction and gene expression data 9-4-2008 Aim To have a mechanism for inferring pathways from gene expression and protein interaction data. Motivation Why
AN EVIDENCE ONTOLOGY FOR USE IN PATHWAY/GENOME DATABASES
 AN EVIDENCE ONTOLOGY FOR USE IN PATHWAY/GENOME DATABASES Appears in Pacific Symposium on Biocomputing 2004 Pages 190-201, World Scientific Publishers, Singapore P.D. KARP, S. PALEY, C.J. KRIEGER SRI International,
AN EVIDENCE ONTOLOGY FOR USE IN PATHWAY/GENOME DATABASES Appears in Pacific Symposium on Biocomputing 2004 Pages 190-201, World Scientific Publishers, Singapore P.D. KARP, S. PALEY, C.J. KRIEGER SRI International,
networks in molecular biology Wolfgang Huber
 networks in molecular biology Wolfgang Huber networks in molecular biology Regulatory networks: components = gene products interactions = regulation of transcription, translation, phosphorylation... Metabolic
networks in molecular biology Wolfgang Huber networks in molecular biology Regulatory networks: components = gene products interactions = regulation of transcription, translation, phosphorylation... Metabolic
Proteomics. Yeast two hybrid. Proteomics - PAGE techniques. Data obtained. What is it?
 Proteomics What is it? Reveal protein interactions Protein profiling in a sample Yeast two hybrid screening High throughput 2D PAGE Automatic analysis of 2D Page Yeast two hybrid Use two mating strains
Proteomics What is it? Reveal protein interactions Protein profiling in a sample Yeast two hybrid screening High throughput 2D PAGE Automatic analysis of 2D Page Yeast two hybrid Use two mating strains
Measuring Semantic Similarity between Gene Ontology Terms
 Measuring Semantic Similarity between Gene Ontology Terms Francisco M. Couto a Mário J. Silva a Pedro M. Coutinho b a Departamento de Informática, Faculdade de Ciências da Universidade de Lisboa, Portugal
Measuring Semantic Similarity between Gene Ontology Terms Francisco M. Couto a Mário J. Silva a Pedro M. Coutinho b a Departamento de Informática, Faculdade de Ciências da Universidade de Lisboa, Portugal
Introduction to clustering methods for gene expression data analysis
 Introduction to clustering methods for gene expression data analysis Giorgio Valentini e-mail: valentini@dsi.unimi.it Outline Levels of analysis of DNA microarray data Clustering methods for functional
Introduction to clustering methods for gene expression data analysis Giorgio Valentini e-mail: valentini@dsi.unimi.it Outline Levels of analysis of DNA microarray data Clustering methods for functional
Probabilistic sparse matrix factorization with an application to discovering gene functions in mouse mrna expression data
 Probabilistic sparse matrix factorization with an application to discovering gene functions in mouse mrna expression data Delbert Dueck, Quaid Morris, and Brendan Frey Department of Electrical and Computer
Probabilistic sparse matrix factorization with an application to discovering gene functions in mouse mrna expression data Delbert Dueck, Quaid Morris, and Brendan Frey Department of Electrical and Computer
Gene expression microarray technology measures the expression levels of thousands of genes. Research Article
 JOURNAL OF COMPUTATIONAL BIOLOGY Volume 7, Number 2, 2 # Mary Ann Liebert, Inc. Pp. 8 DOI:.89/cmb.29.52 Research Article Reducing the Computational Complexity of Information Theoretic Approaches for Reconstructing
JOURNAL OF COMPUTATIONAL BIOLOGY Volume 7, Number 2, 2 # Mary Ann Liebert, Inc. Pp. 8 DOI:.89/cmb.29.52 Research Article Reducing the Computational Complexity of Information Theoretic Approaches for Reconstructing
Identifying Bio-markers for EcoArray
 Identifying Bio-markers for EcoArray Ashish Bhan, Keck Graduate Institute Mustafa Kesir and Mikhail B. Malioutov, Northeastern University February 18, 2010 1 Introduction This problem was presented by
Identifying Bio-markers for EcoArray Ashish Bhan, Keck Graduate Institute Mustafa Kesir and Mikhail B. Malioutov, Northeastern University February 18, 2010 1 Introduction This problem was presented by
Pattern Recognition Letters
 Pattern Recognition Letters 31 (2010) 2133 2137 Contents lists available at ScienceDirect Pattern Recognition Letters journal homepage: www.elsevier.com/locate/patrec Building gene networks with time-delayed
Pattern Recognition Letters 31 (2010) 2133 2137 Contents lists available at ScienceDirect Pattern Recognition Letters journal homepage: www.elsevier.com/locate/patrec Building gene networks with time-delayed
Computational approaches for functional genomics
 Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
Computational approaches for functional genomics Kalin Vetsigian October 31, 2001 The rapidly increasing number of completely sequenced genomes have stimulated the development of new methods for finding
Networks Characteristics Matter for Systems Biology
 Under consideration for publication in Network Science 1 Networks Characteristics Matter for Systems Biology Andrew K. Rider 1,3,4, Tijana Milenković 1,3,4, Geoffrey H. Siwo 2,3,4, Richard S. Pinapati
Under consideration for publication in Network Science 1 Networks Characteristics Matter for Systems Biology Andrew K. Rider 1,3,4, Tijana Milenković 1,3,4, Geoffrey H. Siwo 2,3,4, Richard S. Pinapati
Sparse Factorizations of Gene Expression Guided by Binding Data. L. Badea and D. Tilivea. Pacific Symposium on Biocomputing 10: (2005)
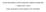 Sparse actorizations of Gene Expression Guided by Binding Data L. Badea and D. ilivea Pacific Symposium on Biocomputing 10:447-458(005) SPARSE ACORIZAIONS O GENE EXPRESSION DAA GUIDED BY BINDING DAA LIVIU
Sparse actorizations of Gene Expression Guided by Binding Data L. Badea and D. ilivea Pacific Symposium on Biocomputing 10:447-458(005) SPARSE ACORIZAIONS O GENE EXPRESSION DAA GUIDED BY BINDING DAA LIVIU
PRINCIPAL COMPONENTS ANALYSIS TO SUMMARIZE MICROARRAY EXPERIMENTS: APPLICATION TO SPORULATION TIME SERIES
 PRINCIPAL COMPONENTS ANALYSIS TO SUMMARIZE MICROARRAY EXPERIMENTS: APPLICATION TO SPORULATION TIME SERIES Soumya Raychaudhuri *, Joshua M. Stuart *, and Russ B. Altman Ψ Stanford Medical Informatics Stanford
PRINCIPAL COMPONENTS ANALYSIS TO SUMMARIZE MICROARRAY EXPERIMENTS: APPLICATION TO SPORULATION TIME SERIES Soumya Raychaudhuri *, Joshua M. Stuart *, and Russ B. Altman Ψ Stanford Medical Informatics Stanford
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 SUMOylation of proteins changes drastically upon heat shock, MG-132 treatment and PR-619 treatment. (a) Schematic overview of all SUMOylation proteins identified to be differentially
Supplementary Figure 1 SUMOylation of proteins changes drastically upon heat shock, MG-132 treatment and PR-619 treatment. (a) Schematic overview of all SUMOylation proteins identified to be differentially
Regulatory Element Detection using a Probabilistic Segmentation Model
 Regulatory Element Detection using a Probabilistic Segmentation Model Harmen J Bussemaker 1, Hao Li 2,3, and Eric D Siggia 2,4 1 Swammerdam Institute for Life Sciences and Amsterdam Center for Computational
Regulatory Element Detection using a Probabilistic Segmentation Model Harmen J Bussemaker 1, Hao Li 2,3, and Eric D Siggia 2,4 1 Swammerdam Institute for Life Sciences and Amsterdam Center for Computational
Gene Ontology. Shifra Ben-Dor. Weizmann Institute of Science
 Gene Ontology Shifra Ben-Dor Weizmann Institute of Science Outline of Session What is GO (Gene Ontology)? What tools do we use to work with it? Combination of GO with other analyses What is Ontology? 1700s
Gene Ontology Shifra Ben-Dor Weizmann Institute of Science Outline of Session What is GO (Gene Ontology)? What tools do we use to work with it? Combination of GO with other analyses What is Ontology? 1700s
Estimating Biological Function Distribution of Yeast Using Gene Expression Data
 Journal of Image and Graphics, Volume 2, No.2, December 2 Estimating Distribution of Yeast Using Gene Expression Data Julie Ann A. Salido Aklan State University, College of Industrial Technology, Kalibo,
Journal of Image and Graphics, Volume 2, No.2, December 2 Estimating Distribution of Yeast Using Gene Expression Data Julie Ann A. Salido Aklan State University, College of Industrial Technology, Kalibo,
Ensemble Non-negative Matrix Factorization Methods for Clustering Protein-Protein Interactions
 Belfield Campus Map Ensemble Non-negative Matrix Factorization Methods for Clustering Protein-Protein Interactions
Belfield Campus Map Ensemble Non-negative Matrix Factorization Methods for Clustering Protein-Protein Interactions
Bayesian Hierarchical Classification. Seminar on Predicting Structured Data Jukka Kohonen
 Bayesian Hierarchical Classification Seminar on Predicting Structured Data Jukka Kohonen 17.4.2008 Overview Intro: The task of hierarchical gene annotation Approach I: SVM/Bayes hybrid Barutcuoglu et al:
Bayesian Hierarchical Classification Seminar on Predicting Structured Data Jukka Kohonen 17.4.2008 Overview Intro: The task of hierarchical gene annotation Approach I: SVM/Bayes hybrid Barutcuoglu et al:
Context dependent visualization of protein function
 Article III Context dependent visualization of protein function In: Juho Rousu, Samuel Kaski and Esko Ukkonen (eds.). Probabilistic Modeling and Machine Learning in Structural and Systems Biology. 2006,
Article III Context dependent visualization of protein function In: Juho Rousu, Samuel Kaski and Esko Ukkonen (eds.). Probabilistic Modeling and Machine Learning in Structural and Systems Biology. 2006,
Introduction to Bioinformatics
 CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
Grouping of correlated feature vectors using treelets
 Grouping of correlated feature vectors using treelets Jing Xiang Department of Machine Learning Carnegie Mellon University Pittsburgh, PA 15213 jingx@cs.cmu.edu Abstract In many applications, features
Grouping of correlated feature vectors using treelets Jing Xiang Department of Machine Learning Carnegie Mellon University Pittsburgh, PA 15213 jingx@cs.cmu.edu Abstract In many applications, features
PROPERTIES OF A SINGULAR VALUE DECOMPOSITION BASED DYNAMICAL MODEL OF GENE EXPRESSION DATA
 Int J Appl Math Comput Sci, 23, Vol 13, No 3, 337 345 PROPERIES OF A SINGULAR VALUE DECOMPOSIION BASED DYNAMICAL MODEL OF GENE EXPRESSION DAA KRZYSZOF SIMEK Institute of Automatic Control, Silesian University
Int J Appl Math Comput Sci, 23, Vol 13, No 3, 337 345 PROPERIES OF A SINGULAR VALUE DECOMPOSIION BASED DYNAMICAL MODEL OF GENE EXPRESSION DAA KRZYSZOF SIMEK Institute of Automatic Control, Silesian University
Clustering gene expression data using graph separators
 1 Clustering gene expression data using graph separators Bangaly Kaba 1, Nicolas Pinet 1, Gaëlle Lelandais 2, Alain Sigayret 1, Anne Berry 1. LIMOS/RR-07-02 13/02/2007 - révisé le 11/05/2007 1 LIMOS, UMR
1 Clustering gene expression data using graph separators Bangaly Kaba 1, Nicolas Pinet 1, Gaëlle Lelandais 2, Alain Sigayret 1, Anne Berry 1. LIMOS/RR-07-02 13/02/2007 - révisé le 11/05/2007 1 LIMOS, UMR
Detection and Visualization of Subspace Cluster Hierarchies
 Detection and Visualization of Subspace Cluster Hierarchies Elke Achtert, Christian Böhm, Hans-Peter Kriegel, Peer Kröger, Ina Müller-Gorman, Arthur Zimek Institute for Informatics, Ludwig-Maimilians-Universität
Detection and Visualization of Subspace Cluster Hierarchies Elke Achtert, Christian Böhm, Hans-Peter Kriegel, Peer Kröger, Ina Müller-Gorman, Arthur Zimek Institute for Informatics, Ludwig-Maimilians-Universität
Uses of the Singular Value Decompositions in Biology
 Uses of the Singular Value Decompositions in Biology The Singular Value Decomposition is a very useful tool deeply rooted in linear algebra. Despite this, SVDs have found their way into many different
Uses of the Singular Value Decompositions in Biology The Singular Value Decomposition is a very useful tool deeply rooted in linear algebra. Despite this, SVDs have found their way into many different
A Study of Network-based Kernel Methods on Protein-Protein Interaction for Protein Functions Prediction
 The Third International Symposium on Optimization and Systems Biology (OSB 09) Zhangjiajie, China, September 20 22, 2009 Copyright 2009 ORSC & APORC, pp. 25 32 A Study of Network-based Kernel Methods on
The Third International Symposium on Optimization and Systems Biology (OSB 09) Zhangjiajie, China, September 20 22, 2009 Copyright 2009 ORSC & APORC, pp. 25 32 A Study of Network-based Kernel Methods on
An Introduction to the spls Package, Version 1.0
 An Introduction to the spls Package, Version 1.0 Dongjun Chung 1, Hyonho Chun 1 and Sündüz Keleş 1,2 1 Department of Statistics, University of Wisconsin Madison, WI 53706. 2 Department of Biostatistics
An Introduction to the spls Package, Version 1.0 Dongjun Chung 1, Hyonho Chun 1 and Sündüz Keleş 1,2 1 Department of Statistics, University of Wisconsin Madison, WI 53706. 2 Department of Biostatistics
10-810: Advanced Algorithms and Models for Computational Biology. Optimal leaf ordering and classification
 10-810: Advanced Algorithms and Models for Computational Biology Optimal leaf ordering and classification Hierarchical clustering As we mentioned, its one of the most popular methods for clustering gene
10-810: Advanced Algorithms and Models for Computational Biology Optimal leaf ordering and classification Hierarchical clustering As we mentioned, its one of the most popular methods for clustering gene
